Continued from EC 1.3.1.51 to EC 1.3.1.126
Sections
EC 1.3.2 With a cytochrome as acceptor
EC 1.3.3 With oxygen as acceptor
EC 1.3.4 With a disulfide as acceptor
EC 1.3.5 With a quinone or related compound as acceptor
EC 1.3.7 With an iron-sulfur protein as acceptor
EC 1.3.8 With flavin as acceptor
EC 1.3.98 With other, known, physiological acceptors
EC 1.3.99 With unknown physiological acceptors
EC 1.3.2.4 fumarate reductase (cytochrome)
[EC 1.3.2.2 Transferred entry: now EC 1.3.99.3 acyl-CoA dehydrogenase (EC 1.3.2.2 created 1961, deleted 1964)]
Accepted name: L-galactonolactone dehydrogenase
Reaction: L-galactono-1,4-lactone + 4 ferricytochrome c = L-dehydroascorbate + 4 ferrocytochrome c + 4 H+ (overall reaction)
(1a) L-galactono-1,4-lactone + 2 ferricytochrome c = L-ascorbate + 2 ferrocytochrome c + 2 H+
(1b) L-ascorbate + 2 ferricytochrome c = L-dehydroascorbate + 2 ferrocytochrome c + 2 H+ (spontaneous)
Other name(s): galactonolactone dehydrogenase; L-galactono-γ-lactone dehydrogenase; L-galactono-γ-lactone:ferricytochrome-c oxidoreductase; GLDHase; GLDase
Systematic name: L-galactono-1,4-lactone:ferricytochrome-c oxidoreductase
Comments: This enzyme catalyses the final step in the biosynthesis of L-ascorbic acid in higher plants and in nearly all higher animals with the exception of primates and some birds [5]. The enzyme is very specific for its substrate L-galactono-1,4-lactone as D-galactono-γ-lactone, D-gulono-γ-lactone, L-gulono-γ-lactone, D-erythronic-γ-lactone, D-xylonic-γ-lactone, L-mannono-γ-lactone, D-galactonate, D-glucuronate and D-gluconate are not substrates [5]. FAD, NAD+, NADP+ and O2 (cf. EC 1.3.3.12, L-galactonolactone oxidase) cannot act as electron acceptor [5].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9029-02-1
References:
1. Mapson, L.W. and Breslow, E. Properties of partially purified L-galactono-γ-lactone dehydrogenase. Biochem. J. 65 (1957) 29 only.
2. Mapson, L.W., Isherwood, F.A. and Chen, Y.T. Biological synthesis of L-ascorbic acid: the conversion of L-galactono-γ-lactone into L-ascorbic acid by plant mitochondria. Biochem. J. 56 (1954) 21-28. [PMID: 13126087]
3. Isherwood, F.A., Chen, Y.T. and Mapson, L.W. Synthesis of L-ascorbic acid in plants and animals. Biochem. J. 56 (1954) 1-15. [PMID: 13126085]
4. Ôba, K., Ishikawa, S., Nishikawa, M., Mizuno, H. and Yamamoto, T. Purification and properties of L-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J. Biochem. (Tokyo) 117 (1995) 120-124. [PMID: 7775377]
5. Østergaard, J., Persiau, G., Davey, M.W., Bauw, G. and Van Montagu, M. Isolation of a cDNA coding for L-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Purification, characterization, cDNA cloning, and expression in yeast. J. Biol. Chem. 272 (1997) 30009-30016. [PMID: 9374475]
Accepted name: fumarate reductase (cytochrome)
Reaction: succinate + 2 ferricytochrome c = fumarate + 2 ferrocytochrome c
Other name(s): fccA (gene name); fcc3 (gene name); flavocytochrome c3
Systematic name: succinate:ferricytochrome-c oxidoreductase
Comments: Contains a non-covalently bound FAD cofactor and four heme c groups. The enzyme, characterized from the bacterium Shewanella frigidimarina, is a soluble periplasmic protein that functions as a terminal electron acceptor during anaerobic growth. The direct electron donor is the membrane-bound tetraheme c-type cytochrome CymA (EC 7.1.1.8, quinol—cytochrome-c reductase), which receives the electrons from the membrane quinol pool.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Pealing, S.L., Black, A.C., Manson, F.D., Ward, F.B., Chapman, S.K. and Reid, G.A. Sequence of the gene encoding flavocytochrome c from Shewanella putrefaciens: a tetraheme flavoenzyme that is a soluble fumarate reductase related to the membrane-bound enzymes from other bacteria. Biochemistry 31 (1992) 12132-12140. [PMID: 1333793]
2. Pealing, S.L., Cheesman, M.R., Reid, G.A., Thomson, A.J., Ward, F.B. and Chapman, S.K. Spectroscopic and kinetic studies of the tetraheme flavocytochrome c from Shewanella putrefaciens NCIMB400. Biochemistry 34 (1995) 6153-6158. [PMID: 7742319]
3. Gordon, E.HJ., Pealing, S.L., Chapman, S.K., Ward, F.B. and Reid, G.A. Physiological function and regulation of flavocytochrome c3, the soluble fumarate reductase from Shewanella putrefaciens NCIMB 400. Microbiology (Reading) 144 (1998) 937-945. [PMID: 9579067]
4. Doherty, M.K., Pealing, S.L., Miles, C.S., Moysey, R., Taylor, P., Walkinshaw, M.D., Reid, G.A. and Chapman, S.K. Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and crystallographic study. Biochemistry 39 (2000) 10695-10701. [PMID: 10978153]
5. Reid, G.A., Miles, C.S., Moysey, R.K., Pankhurst, K.L. and Chapman, S.K. Catalysis in fumarate reductase. Biochim. Biophys Acta 1459 (2000) 310-315. [PMID: 11004445]
6. Schwalb, C., Chapman, S.K. and Reid, G.A. The membrane-bound tetrahaem c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella. Biochem Soc Trans. 30 (2002) 658-662. [PMID: 12196158]
[EC 1.3.3.2 Transferred entry: now EC 1.14.21.6, lathosterol oxidase. NAD(P)H had not been included previously, so enzyme had to be reclassified (EC 1.3.3.2 created 1972, deleted 2005)]
Accepted name: coproporphyrinogen oxidase
Reaction: coproporphyrinogen III + O2 + 2 H+ = protoporphyrinogen-IX + 2 CO2 + 2 H2O
For diagram click here.
Other name(s): coproporphyrinogen III oxidase; coproporphyrinogenase
Systematic name: coproporphyrinogen:oxygen oxidoreductase (decarboxylating)
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9076-84-0
References:
1. Battle, A.M., Benson, A. and Rimington, C. Purification and properties of coproporphyrinogenase. Biochem. J. 97 (1965) 731-740. [PMID: 5881662]
2. Medlock, A.E. and Dailey, H.A. Human coproporphyrinogen oxidase is not a metalloprotein. J. Biol. Chem. 271 (1996) 32507-32510. [PMID: 8955072]
3. Kohno, H., Furukawa, T., Yoshinaga, T., Tokunaga, R. and Taketani, S. Coproporphyrinogen oxidase. Purification, molecular cloning, and induction of mRNA during erythroid differentiation. J. Biol. Chem. 268 (1993) 21359-21363. [PMID: 8407975]
Accepted name: protoporphyrinogen oxidase
Reaction: protoporphyrinogen IX + 3 O2 = protoporphyrin IX + 3 H2O2
For diagram click here.
Other name(s): protoporphyrinogen IX oxidase; protoporphyrinogenase; PPO; Protox; HemG; HemY
Systematic name: protoporphyrinogen-IX:oxygen oxidoreductase
Comments: This is the last common enzyme in the biosynthesis of chlorophylls and heme [8]. Two isoenzymes exist in plants: one in plastids and the other in mitochondria. This is the target enzyme of phthalimide-type and diphenylether-type herbicides [8]. The enzyme from oxygen-dependent species contains FAD [9]. Also slowly oxidizes mesoporphyrinogen IX.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 53986-32-6
References:
1. Poulson, R. The enzymic conversion of protoporphyrinogen IX to protoporphyrin IX in mammalian mitochondria. J. Biol. Chem. 251 (1976) 3730-3733. [PMID: 6461]
2. Poulson, R. and Polglase, W.J. The enzymic conversion of protoporphyrinogen IX to protoporphyrin IX. Protoporphyrinogen oxidase activity in mitochondrial extracts of Saccharomyces cerevisiae. J. Biol. Chem. 250 (1975) 1269-1274. [PMID: 234450]
3. Dailey, H.A. and Dailey, T.A. Protoporphyrinogen oxidase of Myxococcus xanthus. Expression, purification, and characterization of the cloned enzyme. J. Biol. Chem. 271 (1996) 8714-8718. [PMID: 8621504]
4. Wang, K.F., Dailey, T.A. and Dailey, H.A. Expression and characterization of the terminal heme synthetic enzymes from the hyperthermophile Aquifex aeolicus. FEMS Microbiol. Lett. 202 (2001) 115-119. [PMID: 11506917]
5. Corrigall, A.V., Siziba, K.B., Maneli, M.H., Shephard, E.G., Ziman, M., Dailey, T.A., Dailey, H.A., Kirsch. R.E. and Meissner, P.N. Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus subtilis. Arch. Biochem. Biophys. 358 (1998) 251-256. [PMID: 9784236]
6. Ferreira, G.C. and Dailey, H.A. Mouse protoporphyrinogen oxidase. Kinetic parameters and demonstration of inhibition by bilirubin. Biochem. J. 250 (1988) 597–603. [PMID: 2451512]
7. Dailey, T.A. and Dailey, H.A. Human protoporphyrinogen oxidase: expression, purification, and characterization of the cloned enzyme. Protein Sci. 5 (1996) 98–105. [PMID: 8771201]
8. Che, F.S., Watanabe, N., Iwano, M., Inokuchi, H., Takayama, S., Yoshida, S. and Isogai, A. Molecular characterization and subcellular localization of protoporphyrinogen oxidase in spinach chloroplasts. Plant Physiol. 124 (2000) 59–70. [PMID: 10982422]
9. Dailey, T.A. and Dailey, H.A. Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J. Biol. Chem. 273 (1998) 13658–13662. [PMID: 9593705]
Accepted name: bilirubin oxidase
Reaction: 2 bilirubin + O2 = 2 biliverdin + 2 H2O
For diagram click here.
Other name(s): bilirubin oxidase M-1
Systematic name: bilirubin:oxygen oxidoreductase
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 80619-01-8
References:
1. Murao, S. and Tanaka, N. A new enzyme bilirubin oxidase produced by Myrothecium verrucaria MT-1. Agric. Biol. Chem. 45 (1981) 2383-2384.
2. Tanaka, N. and Murao, S. Reaction of bilirubin oxidase produced by Myrothecium verrucaria MT-1. Agr. Biol. Chem. 49 (1985) 843-844.
Accepted name: acyl-CoA oxidase
Reaction: acyl-CoA + O2 = trans-2,3-dehydroacyl-CoA + H2O2
Other name(s): fatty acyl-CoA oxidase; acyl coenzyme A oxidase; fatty acyl-coenzyme A oxidase
Systematic name: acyl-CoA:oxygen 2-oxidoreductase
Comments: A flavoprotein (FAD). Acts on CoA derivatives of fatty acids with chain lengths from 8 to 18.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 61116-22-1
References:
1. Kawaguchi, A., Tsubotani, S., Seyama, Y., Yamakawa, T., Osumi, T., Hashimoto, T., Kikuchi, T., Ando, M. and Okuda, S. Stereochemistry of dehydrogenation catalyzed by acyl-CoA oxidase. J. Biochem. (Tokyo) 88 (1980) 1481-1486. [PMID: 7462191]
2. Osumi, T., Hashimoto, T. and Ui, N. Purification and properties of acyl-CoA oxidase from rat liver. J. Biochem. (Tokyo) 87 (1980) 1735-1746. [PMID: 7400120]
Accepted name: dihydrouracil oxidase
Reaction: 5,6-dihydrouracil + O2 = uracil + H2O2
Systematic name: 5,6-dihydrouracil:oxygen oxidoreductase
Comments: Also oxidizes dihydrothymine to thymine. A flavoprotein (FMN).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 104327-11-9
References:
1. Owaki, J., Uzura, K., Minami, Z. and Kusai, K. Partial-purification and characterization of dihydrouracil oxidase, a flavoprotein from Rhodotorula glutinis. J. Ferment. Technol. 64 (1986) 205-210.
Accepted name: tetrahydroberberine oxidase
Reaction: (S)-tetrahydroberberine + 2 O2 = berberine + 2 H2O2
For diagram click here or here.
Other name(s): (S)-THB oxidase
Systematic name: (S)-tetrahydroberberine:oxygen oxidoreductase
Comments: The enzyme from Berberis sp. (previously listed as EC 1.5.3.8) is a flavoprotein; that from Coptis japonica is not. (R)-Tetrahydroberberines are not oxidized.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 114705-00-9
References:
1. Amann, M., Nagakura, N. and Zenk, M.H. (S)-Tetrahydroprotoberberine oxidase the final enzyme in protoberberine biosynthesis. Tetrahedron Lett. 25 (1984) 953-954.
2. Okada, N., Shinmyo, A., Okada, H. and Yamada, Y. Purification and characterization of (S)-tetrahydroberberine oxidase from cultured Coptis japonica cells. Phytochemistry 27 (1988) 979-982.
[EC 1.3.3.9 Transferred entry: secologanin synthase. Now EC 1.14.19.62, secologanin synthase (EC 1.3.3.9 created 2002, deleted 2018)]
Accepted name: tryptophan α,β-oxidase
Reaction: L-tryptophan + O2 = α,β-didehydrotryptophan + H2O2
Other name(s): L-tryptophan 2',3'-oxidase; L-tryptophan α,β-dehydrogenase
Systematic name: L-tryptophan:oxygen α,β-oxidoreductase
Comments: Requires heme. The enzyme from Chromobacterium violaceum is specific for tryptophan derivatives possessing its carboxyl group free or as an amide or ester, and an unsubstituted indole ring. Also catalyses the α,β dehydrogenation of L-tryptophan side chains in peptides. The product of the reaction can hydrolyse spontaneously to form indolepyruvate.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 156859-19-7
References:
1. Genet, R., Denoyelle, C. and Menez, A. Purification and partial characterization of an amino acid α,β-dehydrogenase, L-tryptophan 2',3'-oxidase from Chromobacterium violaceum. J. Biol. Chem. 269 (1994) 18177-18184. [PMID: 8027079]
2. Genet, R., Benetti, P.H., Hammadi, A. and Menez, A. L-Tryptophan 2',3'-oxidase from Chromobacterium violaceum. Substrate specificity and mechanistic implications. J. Biol. Chem. 270 (1995) 23540-23545. [PMID: 7559518]
Accepted name: pyrroloquinoline-quinone synthase
Reaction: 6-(2-amino-2-carboxyethyl)-7,8-dioxo-1,2,3,4,7,8-hexahydroquinoline-2,4-dicarboxylate + 3 O2 = 4,5-dioxo-1H-pyrrolo[2,3-f]quinoline-2,7,9-tricarboxylate + 2 H2O2 + 2 H2O
For diagram click here.
Other name(s): PqqC
Systematic name: 6-(2-amino-2-carboxyethyl)-7,8-dioxo-1,2,3,4,5,6,7,8-octahydroquinoline-2,4-dicarboxylate:oxygen oxidoreductase (cyclizing)
Comments: So far only a single turnover of the enzyme has been observed, and the pyrroloquinoline quinone remains bound to it. It is not yet known what releases the product in the bacterium.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 353484-42-1
References:
1. Magnusson, O.T., Toyama, H., Saeki, M., Schwarzenbacher, R. and Klinman, J.P. The structure of a biosynthetic intermediate of pyrroloquinoline quinone (PQQ) and elucidation of the final step of PQQ biosynthesis. J. Am. Chem. Soc. 126 (2004) 5342-5343. [PMID: 15113189]
2. Magnusson, O.T., Toyama, H., Saeki, M., Rojas, A., Reed, J.C., Liddington, R.C., Klinman, J.P. and Schwarzenbacher, R. Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl. Acad. Sci. USA 101 (2004) 7913-7918. [PMID: 15148379]
3. Toyama, H., Chistoserdova, L. and Lidstrom, M.E. Sequence analysis of pqq genes required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1 and the purification of a biosynthetic intermediate. Microbiology 143 (1997) 595-602. [PMID: 9043136]
4. Toyama, H., Fukumoto, H., Saeki, M., Matsushita, K., Adachi, O. and Lidstrom, M.E. PqqC/D, which converts a biosynthetic intermediate to pyrroloquinoline quinone. Biochem. Biophys. Res. Commun. 299 (2002) 268-272. [PMID: 1243798]
5. Schwarzenbacher, R., Stenner-Liewen, F., Liewen, H., Reed, J.C. and Liddington, R.C. Crystal structure of PqqC from Klebsiella pneumoniae at 2.1 Å resolution. Protein 56 (2004) 401-403. [PMID: 15211525]
Accepted name: L-galactonolactone oxidase
Reaction: L-galactono-1,4-lactone + O2 = L-ascorbate + H2O2
Other name(s): L-galactono-1,4-lactone oxidase
Systematic name: L-galactono-1,4-lactone:oxygen 3-oxidoreductase
Comments: A flavoprotein. Acts on the 1,4-lactones of L-galactonic, D-altronic, L-fuconic, D-arabinic and D-threonic acids; not identical with EC 1.1.3.8 L-gulonolactone oxidase. (cf. EC 1.3.2.3 galactonolactone dehydrogenase).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Bleeg, H.S. and Christensen, F. Biosynthesis of ascorbate in yeast. Purification of L-galactono-1,4-lactone oxidase with properties different from mammalian L-gulonolactone oxidase. Eur. J. Biochem. 127 (1982) 391-396. [PMID: 6754380]
Accepted name: albonoursin synthase
Reaction: cyclo(L-leucyl-L-phenylalanyl) + 2 O2 = albonoursin + 2 H2O2 (overall reaction)
(1a) cyclo(L-leucyl-L-phenylalanyl) + O2 = cyclo[(Z)-α,β-didehydrophenylalanyl-L-leucyl] + H2O2
(1b) cyclo[(Z)-α,β-didehydrophenylalanyl-L-leucyl] + O2 = albonoursin + H2O2
For diagram of reaction click here.
Glossary: cyclo(L-leucyl-L-phenylalanyl) = (3S,6S)-3-benzyl-6-(2-methylpropyl)piperazine-2,5-dione
cyclo[(Z)-α,β-didehydrophenylalanyl-L-leucyl] = (3Z,6S)-3-benzylidene-6-(2-methylpropyl)piperazine-2,5-dione
albonoursin = (3Z,6Z)-3-benzylidene-6-(2-methylpropylidene)piperazine-2,5-dione
Other name(s): cyclo(dipeptide):oxygen oxidoreductase; cyclic dipeptide oxidase; AlbA
Systematic name: cyclo(L-leucyl-L-phenylalanyl):oxygen oxidoreductase
Comments: A flavoprotein from the bacterium Streptomyces noursei. The enzyme can also oxidize several other cyclo dipeptides, the best being cyclo(L-tryptophyl-L-tryptophyl) and cyclo(L-phenylalanyl-L-phenylalanyl) [1,2].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Gondry, M., Lautru, S., Fusai, G., Meunier, G., Menez, A. and Genet, R. Cyclic dipeptide oxidase from Streptomyces noursei. Isolation, purification and partial characterization of a novel, amino acyl α,β-dehydrogenase. Eur. J. Biochem. 268 (2001) 1712-1721. [PMID: 11248691]
2. Lautru, S., Gondry, M., Genet, R. and Pernodet, J.L. The albonoursin gene cluster of S. noursei. Biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 9 (2002) 1355-1364. [PMID: 12498889]
Accepted name: aclacinomycin-A oxidase
Reaction: aclacinomycin A + O2 = aclacinomycin Y + H2O2
For diagram of reaction click here.
Glossary: aclacinomycin A = 2-ethyl-1,2,3,4,6,11-hexahydro-2,5,7-trihydroxy-6,11-dioxo-4-[[2,3,6-trideoxy-4-O-[2,6-dideoxy-4-O-[(2R,6S)-tetrahydro-6-methyl-5-oxo-2H-pyran-2-yl]-α-L-lyxo-hexopyranosyl]-3-(dimethylamino)-α-L-lyxo-hexopyranosyl]oxy]naphthacene-1-carboxylic acid methyl ester
aclacinomycin Y = 2-ethyl-1,2,3,4,6,11-hexahydro-2,5,7-trihydroxy-6,11-dioxo-4-[[2,3,6-trideoxy-4-O-[2,6-dideoxy-4-O-[(2R,6S)-5,6-dihydro-6-methyl-5-oxo-2H-pyran-2-yl]-α-L-lyxo-hexopyranosyl]-3-(dimethylamino)-α-L-lyxo-hexopyranosyl]oxy]naphthacene-1-carboxylic acid methyl ester
Other name(s): AknOx (ambiguous); aclacinomycin oxidoreductase (ambiguous)
Systematic name: aclacinomycin-A:oxygen oxidoreductase
Comments: A flavoprotein (FAD). This bifunctional enzyme is a secreted flavin-dependent enzyme that is involved in the modification of the terminal sugar residues in the biosynthesis of aclacinomycins. The enzyme utilizes the same active site to catalyse the oxidation of the rhodinose moiety of aclacinomycin N to the cinerulose A moiety of aclacinomycin A (cf. EC 1.1.3.45) and the oxidation of the latter to the L-aculose moiety of aclacinomycin Y.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Yoshimoto, A., Ogasawara, T., Kitamura, I., Oki, T., Inui, T., Takeuchi, T. and Umezawa, H. Enzymatic conversion of aclacinomycin A to Y by a specific oxidoreductase in Streptomyces. J. Antibiot. (Tokyo) 32 (1979) 472-481. [PMID: 528393]
2. Alexeev, I., Sultana, A., Mantsala, P., Niemi, J. and Schneider, G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl. Acad. Sci. USA 104 (2007) 6170-6175. [PMID: 17395717]
3. Sultana, A., Alexeev, I., Kursula, I., Mantsala, P., Niemi, J. and Schneider, G. Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning. Acta Crystallogr. D Biol. Crystallogr. 63 (2007) 149-159. [PMID: 17242508]
Accepted name: coproporphyrinogen III oxidase (coproporphyrin-forming)
Reaction: coproporphyrinogen III + 3 O2 = coproporphyrin III + 3 H2O2
Other name(s): hemY (gene name)
Systematic name: coproporphyrinogen-III:oxygen oxidoreductase (coproporphyrin-forming)
Comments: Contains FAD. The enzyme, present in Gram-positive bacteria, participates in heme biosynthesis. It can also catalyse the reaction of EC 1.3.3.4, protoporphyrinogen oxidase, at a lower level.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Hansson, M. and Hederstedt, L. Bacillus subtilis HemY is a peripheral membrane protein essential for protoheme IX synthesis which can oxidize coproporphyrinogen III and protoporphyrinogen IX. J. Bacteriol. 176 (1994) 5962-5970. [PMID: 7928957]
2. Corrigall, A.V., Siziba, K.B., Maneli, M.H., Shephard, E.G., Ziman, M., Dailey, T.A., Dailey, H.A., Kirsch, R.E. and Meissner, P.N. Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus subtilis. Arch. Biochem. Biophys. 358 (1998) 251-256. [PMID: 9784236]
3. Qin, X., Sun, L., Wen, X., Yang, X., Tan, Y., Jin, H., Cao, Q., Zhou, W., Xi, Z. and Shen, Y. Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J. Struct. Biol. 170 (2010) 76-82. [PMID: 19944166]
4. Dailey, H.A., Gerdes, S., Dailey, T.A., Burch, J.S. and Phillips, J.D. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. USA 112 (2015) 2210-2215. [PMID: 25646457]
Accepted name: oxazoline dehydrogenase
Reaction: (1) a [protein]-(1S,4R)-2-(C-substituted-aminomethyl)-4-acyl-2-thiazoline + O2 = a [protein]-(S)-2-(C-substituted-aminomethyl)-4-acyl-1,3-thiazole + H2O2
(2) a [protein]-(S,S)-2-(C-substituted-aminomethyl)-4-acyl-2-oxazoline + O2 = a [protein]-(S)-2-(C-substituted-aminomethyl)-4-acyl-1,3-oxazole + H2O2
(3) a [protein]-(S,S)-2-(C-substituted-aminomethyl)-4-acyl-5-methyl-2-oxazoline + O2 = a [protein]-(S)-2-(C-substituted-aminomethyl)-4-acyl-5-methyl-1,3-oxazole + H2O2
Other name(s): azoline oxidase; thiazoline oxidase; cyanobactin oxidase; patG (gene name); mcaG (gene name); artG (gene name); lynG (gene name); tenG (gene name)
Systematic name: a [protein]-2-oxazoline:oxygen oxidoreductase (2-oxazole-forming)
Comments: Contains FMN. This enzyme oxidizes 2-oxazoline, 5-methyl-2-oxazoline, and 2-thiazoline within peptides, which were formed by EC 6.2.2.2, oxazoline synthase, and EC 6.2.2.3, thiazoline synthase, to the respective pyrrole-type rings. The enzyme is found as either a stand-alone protein or as a domain within a multifunctional protein (the G protein) that also functions as a peptidase.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Li, Y.M., Milne, J.C., Madison, L.L., Kolter, R. and Walsh, C.T. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274 (1996) 1188-1193. [PMID: 8895467]
2. Schmidt, E.W., Nelson, J.T., Rasko, D.A., Sudek, S., Eisen, J.A., Haygood, M.G. and Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102 (2005) 7315-7320. [PMID: 15883371]
3. Bent, A.F., Mann, G., Houssen, W.E., Mykhaylyk, V., Duman, R., Thomas, L., Jaspars, M., Wagner, A. and Naismith, J.H. Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD. Acta Crystallogr D Struct Biol 72 (2016) 1174-1180. [PMID: 27841750]
4. Ghilarov, D., Stevenson, C.EM., Travin, D.Y., Piskunova, J., Serebryakova, M., Maxwell, A., Lawson, D.M. and Severinov, K. Architecture of microcin B17 synthetase: an octameric protein complex converting a ribosomally synthesized peptide into a DNA gyrase poison. Mol. Cell 73 (2019) 749-762.e5. [PMID: 30661981]
Accepted name: benzylmalonyl-CoA dehydrogenase
Reaction: benzylmalonyl-CoA + O2 = (E)-cinnamoyl-CoA + CO2 + H2O2
Other name(s): iaaF (gene name)
Systematic name: benzylmalonyl-CoA:oxygen oxidoreductase (decarboxylating)
Comments: The enzyme, characterized from the bacterium Aromatoleum aromaticum, is involved in degradation of (indol-3-yl)acetate, where it is believed to function on (2-aminobenzyl)malonyl-CoA.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Schuhle, K., Saft, M., Vogeli, B., Erb, T.J. and Heider, J. Benzylmalonyl-CoA dehydrogenase, an enzyme involved in bacterial auxin degradation. Arch. Microbiol. 203 (2021) 4149-4159. [PMID: 34059946]
Accepted name: fumarate reductase (CoM/CoB)
Reaction: fumarate + CoM + CoB = succinate + CoM-S-S-CoB
Glossary: CoB = coenzyme B = N-(7-mercaptoheptanoyl)threonine 3-O-phosphate = N-(7-thioheptanoyl)-3-O-phosphothreonine
CoM = coenzyme M = 2-sulfanylethane-1-sulfonate = 2-mercaptoethanesulfonate (deprecated)
Other name(s): thiol:fumarate reductase; Tfr
Systematic name: fumarate CoM:CoB oxidoreductase (succinate-forming)
Comments: The enzyme, isolated from the archaeon Methanobacterium thermoautotrophicum, is very oxygen sensitive. It cannot use reduced flavins, reduced coenzyme F420, or NAD(P)H as an electron donor. Distinct from EC 1.3.1.6 [fumarate reductase (NADH)], EC 1.3.5.1 [succinate dehydrogenase (ubiquinone)], and EC 1.3.5.4 [fumarate reductase (quinol)].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Khandekar, S.S. and Eirich, L.D. Purification and characterization of an anabolic fumarate reductase from Methanobacterium thermoautotrophicum. Appl. Environ. Microbiol. 55 (1989) 856-861. [PMID: 2499256]
2. Heim, S., Kunkel, A., Thauer, R.K. and Hedderich, R. Thiol:fumarate reductase (Tfr) from Methanobacterium thermoautotrophicum. Identification of the catalytic sites for fumarate reduction and thiol oxidation. Eur. J. Biochem. 253 (1998) 292-299. [PMID: 9578488]
EC 1.3.5.1 succinate dehydrogenase
EC 1.3.5.2 dihydroorotate dehydrogenase (quinone)
EC 1.3.5.3 protoporphyrinogen IX dehydrogenase (quinone)
EC 1.3.5.4 now included in EC 1.3.5.1
EC 1.3.5.5 15-cis-phytoene desaturase
EC 1.3.5.6 9,9'-dicis-ζ-carotene desaturase
EC 1.3.5.1
Accepted name: succinate dehydrogenase
Reaction: succinate + a quinone = fumarate + a quinol
For diagram of reaction, click here
Other name(s): succinate dehydrogenase (quinone); succinate dehydrogenase (ubiquinone); succinic dehydrogenase; complex II (ambiguous); succinate dehydrogenase complex; SDH (ambiguous); succinate:ubiquinone oxidoreductase; fumarate reductase (quinol); FRD; menaquinol-fumarate oxidoreductase; succinate dehydrogenase (menaquinone); succinate:menaquinone oxidoreductase; fumarate reductase (menaquinone)
Systematic name: succinate:quinone oxidoreductase
Comments: A complex generally comprising an FAD-containing component that also binds the carboxylate substrate (A subunit), a component that contains three different iron-sulfur centers [2Fe-2S], [4Fe-4S], and [3Fe-4S] (B subunit), and a hydrophobic membrane-anchor component (C, or C and D subunits) that is also the site of the interaction with quinones. The enzyme is found in the inner mitochondrial membrane in eukaryotes and the plasma membrane of bacteria and archaea, with the hydrophilic domain extending into the mitochondrial matrix and the cytoplasm, respectively. Under aerobic conditions the enzyme catalyses succinate oxidation, a key step in the citric acid (TCA) cycle, transferring the electrons to quinones in the membrane, thus linking the TCA cycle with the aerobic respiratory chain (where it is known as complex II). Under anaerobic conditions the enzyme functions as a fumarate reductase, transferring electrons from the quinol pool to fumarate, and participating in anaerobic respiration with fumarate as the terminal electron acceptor. The enzyme interacts with the quinone produced by the organism, such as ubiquinone, menaquinone, caldariellaquinone, thermoplasmaquinone, rhodoquinone etc. Some of the enzymes contain two heme subunits in their membrane anchor subunit. These enzymes catalyse an electrogenic reaction and are thus classified as EC 7.1.1.12, succinate dehydrogenase (electrogenic, proton-motive force generating).
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number: 9028-11-9
References:
1. Kita, K., Vibat, C.R., Meinhardt, S., Guest, J.R. and Gennis, R.B. One-step purification from Escherichia coli of complex II (succinate: ubiquinone oxidoreductase) associated with succinate-reducible cytochrome b556. J. Biol. Chem. 264 (1989) 2672-2677. [PMID: 2644269]
2. Van Hellemond, J.J. and Tielens, A.G. Expression and functional properties of fumarate reductase. Biochem. J. 304 (1994) 321-331. [PMID: 7998964]
3. Iverson, T.M., Luna-Chavez, C., Cecchini, G. and Rees, D.C. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284 (1999) 1961-1966. [PMID: 10373108]
4. Cecchini, G., Schroder, I., Gunsalus, R.P. and Maklashina, E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553 (2002) 140-157. [PMID: 11803023]
5. Figueroa, P., Leon, G., Elorza, A., Holuigue, L., Araya, A. and Jordana, X. The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol. Biol. 50 (2002) 725-734. [PMID: 12374303]
6. Cecchini, G. Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72 (2003) 77-109. [PMID: 14527321]
7. Oyedotun, K.S. and Lemire, B.D. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J. Biol. Chem. 279 (2004) 9424-9431. [PMID: 14672929]
8. Kurokawa, T. and Sakamoto, J. Purification and characterization of succinate:menaquinone oxidoreductase from Corynebacterium glutamicum. Arch. Microbiol. 183 (2005) 317-324. [PMID: 15883782]
9. Iwata, F., Shinjyo, N., Amino, H., Sakamoto, K., Islam, M.K., Tsuji, N. and Kita, K. Change of subunit composition of mitochondrial complex II (succinate-ubiquinone reductase/quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol Int 57 (2008) 54-61. [PMID: 17933581]
Accepted name: dihydroorotate dehydrogenase (quinone)
Reaction: (S)-dihydroorotate + a quinone = orotate + a quinol
Other name(s): dihydroorotate:ubiquinone oxidoreductase; (S)-dihydroorotate:(acceptor) oxidoreductase; (S)-dihydroorotate:acceptor oxidoreductase; DHOdehase (ambiguous); DHOD (ambiguous); DHODase (ambiguous); DHODH
Systematic name: (S)-dihydroorotate:quinone oxidoreductase
Comments: This Class 2 dihydroorotate dehydrogenase enzyme contains FMN [4]. The enzyme is found in eukaryotes in the mitochondrial membrane, and in some Gram negative bacteria associated with the cytoplasmic membrane [2,5]. The reaction is the only redox reaction in the de-novo biosynthesis of pyrimidine nucleotides [2,4]. The best quinone electron acceptors for the enzyme from bovine liver are ubiquinone-6 and ubiquinone-7, although simple quinones, such as benzoquinone, can also act as acceptor at lower rates [2]. Methyl-, ethyl-, tert-butyl and benzyl-(S)-dihydroorotates are also substrates, but 1- and 3-methyl and 1,3-dimethyl methyl-(S)-dihydroorotates are not [2]. Class 1 dihydroorotate dehydrogenases use either fumarate (EC 1.3.98.1), NAD+ (EC 1.3.1.14) or NADP+ (EC 1.3.1.15) as electron acceptor.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 59088-23-2
References:
1. Forman, H.J. and Kennedy, J. Mammalian dihydroorotate dehydrogenase: physical and catalytic properties of the primary enzyme. Arch. Biochem. Biophys. 191 (1978) 23-31. [PMID: 216313]
2. Hines, V., Keys, L.D., III and Johnston, M. Purification and properties of the bovine liver mitochondrial dihydroorotate dehydrogenase. J. Biol. Chem. 261 (1986) 11386-11392. [PMID: 3733756]
3. Bader, B., Knecht, W., Fries, M. and Löffler, M. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr. Purif. 13 (1998) 414-422. [PMID: 9693067]
4. Fagan, R.L., Nelson, M.N., Pagano, P.M. and Palfey, B.A. Mechanism of flavin reduction in Class 2 dihydroorotate dehydrogenases. Biochemistry 45 (2006) 14926-14932. [PMID: 17154530]
5. Björnberg, O., Grüner, A.C., Roepstorff, P. and Jensen, K.F. The activity of Escherichia coli dihydroorotate dehydrogenase is dependent on a conserved loop identified by sequence homology, mutagenesis, and limited proteolysis. Biochemistry 38 (1999) 2899-2908. [PMID: 10074342]
Accepted name: protoporphyrinogen IX dehydrogenase (quinone)
Reaction: protoporphyrinogen IX + 3 quinone = protoporphyrin IX + 3 quinol
Other name(s): HemG; protoporphyrinogen IX dehydrogenase (menaquinone)
Systematic name: protoporphyrinogen IX:quinone oxidoreductase
Comments: Contains FMN. The enzyme participates in heme b biosynthesis. In the bacterium Escherichia coli it interacts with either ubiquinone or menaquinone, depending on whether the organism grows aerobically or anaerobically.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Boynton, T.O., Daugherty, L.E., Dailey, T.A. and Dailey, H.A. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48 (2009) 6705-6711. [PMID: 19583219]
2. Möbius, K., Arias-Cartin, R., Breckau, D., Hännig, A.L., Riedmann, K., Biedendieck, R., Schroder, S., Becher, D., Magalon, A., Moser, J., Jahn, M. and Jahn, D. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. USA 107 (2010) 10436-10441. [PMID: 20484676]
[EC 1.3.5.4 Transferred entry: fumarate reductase (quinol), now included in EC 1.3.5.1, succinate dehydrogenase. (EC 1.3.5.4 created 2010, modified 2013, deleted 2022)]
Accepted name: 15-cis-phytoene desaturase
Reaction: 15-cis-phytoene + 2 plastoquinone = 9,15,9'-tricis-ζ-carotene + 2 plastoquinol (overall reaction)
(1a) 15-cis-phytoene + plastoquinone = 15,9'-dicis-phytofluene + plastoquinol
(1b) 15,9'-dicis-phytofluene + plastoquinone = 9,15,9'-tricis-ζ-carotene + plastoquinol
For diagram of reaction click here.
Other name(s): phytoene desaturase (ambiguous); PDS; plant-type phytoene desaturase
Systematic name: 15-cis-phytoene:plastoquinone oxidoreductase
Comments: This enzyme is involved in carotenoid biosynthesis in plants and cyanobacteria. The enzyme from Synechococcus can also use NAD+ and NADP+ as electron acceptor under anaerobic conditions. The enzyme from Gentiana lutea shows no activity with NAD+ or NADP+ [1].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Breitenbach, J., Zhu, C. and Sandmann, G. Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. J. Agric. Food Chem. 49 (2001) 5270-5272. [PMID: 11714315]
2. Schneider, C., Boger, P. and Sandmann, G. Phytoene desaturase: heterologous expression in an active state, purification, and biochemical properties. Protein Expr. Purif. 10 (1997) 175-179. [PMID: 9226712]
3. Fraser, P.D., Linden, H. and Sandmann, G. Purification and reactivation of recombinant Synechococcus phytoene desaturase from an overexpressing strain of Escherichia coli. Biochem. J. 291 (1993) 687-692. [PMID: 8489496]
4. Breitenbach, J. and Sandmann, G. ζ-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220 (2005) 785-793. [PMID: 15503129]
Accepted name: 9,9'-dicis-ζ-carotene desaturase
Reaction: 9,9'-dicis-ζ-carotene + 2 quinone = 7,9,7',9'-tetracis-lycopene + 2 quinol (overall reaction)
(1a) 9,9'-dicis-ζ-carotene + a quinone = 7,9,9'-tricis-neurosporene + a quinol
(1b) 7,9,9'-tricis-neurosporene + a quinone = 7,9,7',9'-tetracis-lycopene + a quinol
For diagram of reaction click here.
Glossary: 7,9,7',9'-tetracis-lycopene = prolycopene
Other name(s): ζ-carotene desaturase; ZDS
Systematic name: 9,9'-dicis-ζ-corotene:quinone oxidoreductase
Comments: This enzyme is involved in carotenoid biosynthesis in plants and cyanobacteria.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Albrecht, M., Linden, H. and Sandmann, G. Biochemical characterization of purified ζ-carotene desaturase from Anabaena PCC 7120 after expression in E. coli. Eur. J. Biochem. 236 (1996) 115-120. [PMID: 8617254]
2. Josse, E.M., Simkin, A.J., Gaffe, J., Laboure, A.M., Kuntz, M. and Carol, P. A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 123 (2000) 1427-1436. [PMID: 10938359]
3. Breitenbach, J., Kuntz, M., Takaichi, S. and Sandmann, G. Catalytic properties of an expressed and purified higher plant type ζ-carotene desaturase from Capsicum annuum. Eur. J. Biochem. 265 (1999) 376-383. [PMID: 10491195]
4. Breitenbach, J. and Sandmann, G. ζ-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220 (2005) 785-793. [PMID: 15503129]
Accepted name: 6-hydroxynicotinate reductase
Reaction: 6-oxo-1,4,5,6-tetrahydronicotinate + oxidized ferredoxin = 6-hydroxynicotinate + reduced ferredoxin
For diagram click here.
Other name(s): 6-oxotetrahydronicotinate dehydrogenase; 6-hydroxynicotinic reductase; HNA reductase
Systematic name: 6-oxo-1,4,5,6-tetrahydronicotinate:ferredoxin oxidoreductase
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 9030-84-6
References:
1. Holcenberg, J.S. and Tsai, L. Nicotinic acid metabolism. IV. Ferredoxin-dependent reduction of 6-hydroxynicotinic acid to 6-oxo-1,4,5,6-tetrahydronicotinic acid. J. Biol. Chem. 244 (1969) 1204-1211. [PMID: 5767303]
Accepted name: 15,16-dihydrobiliverdin:ferredoxin oxidoreductase
Reaction: 15,16-dihydrobiliverdin + oxidized ferredoxin = biliverdin IXα + reduced ferredoxin
For diagram click here.
Other name(s): PebA
Systematic name: 15,16-dihydrobiliverdin:ferredoxin oxidoreductase
Comments: Catalyses the two-electron reduction of biliverdin IXα at the C15 methine bridge. It has been proposed that this enzyme and EC 1.3.7.3, phycoerythrobilin:ferredoxin oxidoreductase, function as a dual enzyme complex in the conversion of biliverdin IXα into phycoerythrobilin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 347401-20-1
References:
1. Frankenberg, N., Mukougawa, K., Kohchi, T. and Lagarias, J.C. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13 (2001) 965-978. [PMID: 11283349]
Accepted name: phycoerythrobilin:ferredoxin oxidoreductase
Reaction: (3Z)-phycoerythrobilin + oxidized ferredoxin = 15,16-dihydrobiliverdin + reduced ferredoxin
For diagram click here.
Other name(s): PebB
Systematic name: (3Z)-phycoerythrobilin:ferredoxin oxidoreductase
Comments: Catalyses the two-electron reduction of the C2 and C31 diene system of 15,16-dihydrobiliverdin. Specific for 15,16-dihydrobiliverdin. It has been proposed that this enzyme and EC 1.3.7.2, 15,16-dihydrobiliverdin:ferredoxin oxidoreductase, function as a dual enzyme complex in the conversion of biliverdin IXα to phycoerythrobilin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 347401-21-2
References:
1. Frankenberg, N., Mukougawa, K., Kohchi, T. and Lagarias, J.C. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13 (2001) 965-978. [PMID: 11283349]
Accepted name: phytochromobilin:ferredoxin oxidoreductase
Reaction: (3Z)-phytochromobilin + oxidized ferredoxin = biliverdin IXα + reduced ferredoxin
For diagram click here.
Other name(s): HY2; PΦB synthase; phytochromobilin synthase
Systematic name: (3Z)-phytochromobilin:ferredoxin oxidoreductase
Comments: Catalyses the two-electron reduction of biliverdin IXα. Can use [2Fe-2S] ferredoxins from a number of sources as acceptor but not the [4Fe-4S] ferredoxin from Clostridium pasteurianum. The isomerization of (3Z)-phytochromobilin to (3E)-phytochromobilin is thought to occur prior to covalent attachment to apophytochrome in the plant cell cytoplasm. Flavodoxins can be used instead of ferredoxin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 138263-99-7
References:
1. Frankenberg, N., Mukougawa, K., Kohchi, T. and Lagarias, J.C. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13 (2001) 965-978. [PMID: 11283349]
2. McDowell, M.T. and Lagarias, J.C. Purification and biochemical properties of phytochromobilin synthase from etiolated oat seedlings. Plant Physiol. 126 (2001) 1546-1554. [PMID: 11500553]
3. Terry, M.J., Wahleithner, J.A. and Lagarias, J.C. Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306 (1993) 1-15. [PMID: 8215388]
Accepted name: phycocyanobilin:ferredoxin oxidoreductase
Reaction: (3Z)-phycocyanobilin + 4 oxidized ferredoxin = biliverdin IXα + 4 reduced ferredoxin
For diagram of reaction click here.
Systematic name: (3Z)-phycocyanobilin:ferredoxin oxidoreductase
Comments: Catalyses the four-electron reduction of biliverdin IXα (2-electron reduction at both the A and D rings). Reaction proceeds via an isolatable 2-electron intermediate, 181,182-dihydrobiliverdin. Flavodoxins can be used instead of ferredoxin. The direct conversion of biliverdin IXα (BV) to (3Z)-phycocyanolbilin (PCB) in the cyanobacteria Synechocystis sp. PCC 6803, Anabaena sp. PCC7120 and Nostoc punctiforme is in contrast to the proposed pathways of PCB biosynthesis in the red alga Cyanidium caldarium, which involves (3Z)-phycoerythrobilin (PEB) as an intermediate [2] and in the green alga Mesotaenium caldariorum, in which PCB is an isolable intermediate.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number: 347401-12-1
References:
1. Frankenberg, N., Mukougawa, K., Kohchi, T. and Lagarias, J.C. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13 (2001) 965-978. [PMID: 11283349]
2. Beale, S.I. Biosynthesis of phycobilins. Chem. Rev. 93 (1993) 785-802.
3. Wu, S.-H., McDowell, M.T. and Lagarias, J.C. Phycocyanobilin is the natural chromophore precursor of phytochrome from the green alga Mesotaenium caldariorum. J. Biol. Chem. 272 (1997) 25700-25705. [PMID: 9325294]
Accepted name: phycoerythrobilin synthase
Reaction: (3Z)-phycoerythrobilin + 2 oxidized ferredoxin = biliverdin IXα + 2 reduced ferredoxin
Other name(s): PebS
Systematic name: (3Z)-phycoerythrobilin:ferredoxin oxidoreductase (from biliverdin IXα)
Comments: This enzyme, from a cyanophage infecting oceanic cyanobacteria of the Prochlorococcus genus, uses a four-electron reduction to carry out the reactions catalysed by EC 1.3.7.2 (15,16-dihydrobiliverdin:ferredoxin oxidoreductase) and EC 1.3.7.3 (phycoerythrobilin:ferredoxin oxidoreductase). 15,16-Dihydrobiliverdin is formed as a bound intermediate. Free 15,16-dihydrobiliverdin can also act as a substrate to form phycoerythrobilin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Dammeyer, T., Bagby, S.C., Sullivan, M.B., Chisholm, S.W. and Frankenberg-Dinkel, N. Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr. Biol. 18 (2008) 442-448. [PMID: 18356052]
Accepted name: ferredoxin:protochlorophyllide reductase (ATP-dependent)
Reaction: chlorophyllide a + oxidized ferredoxin + 2 ADP + 2 phosphate = protochlorophyllide a + reduced ferredoxin + 2 ATP + 2 H2O
For diagram of reaction click here.
Other name(s): light-independent protochlorophyllide reductase
Systematic name: ATP-dependent ferredoxin:protochlorophyllide-a 7,8-oxidoreductase
Comments: Occurs in photosynthetic bacteria, cyanobacteria, green algae and gymnosperms. The enzyme catalyses trans-reduction of the D-ring of protochlorophyllide; the product has the (7S,8S)-configuration. Unlike EC 1.3.1.33 (protochlorophyllide reductase), light is not required. The enzyme contains a [4Fe-4S] cluster, and structurally resembles the Fe protein/MoFe protein complex of nitrogenase (EC 1.18.6.1), which catalyses an ATP-driven reduction.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Fujita, Y., Matsumoto, H., Takahashi, Y. and Matsubara, H. Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 34 (1993) 305-314. [PMID: 8199775]
2. Nomata, J., Ogawa, T., Kitashima, M., Inoue, K. and Fujita, Y. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 582 (2008) 1346-1350. [PMID: 18358835]
3. Muraki, N., Nomata, J., Ebata, K., Mizoguchi, T., Shiba, T., Tamiaki, H., Kurisu, G. and Fujita, Y. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465 (2010) 110-114. [PMID: 20400946]
Accepted name: benzoyl-CoA reductase
Reaction: cyclohexa-1,5-diene-1-carbonyl-CoA + oxidized ferredoxin + 2 ADP + 2 phosphate = benzoyl-CoA + reduced ferredoxin + 2 ATP + 2 H2O
For diagram of reaction click here.
Other name(s): benzoyl-CoA reductase (dearomatizing)
Systematic name: cyclohexa-1,5-diene-1-carbonyl-CoA:ferredoxin oxidoreductase (aromatizing, ATP-forming)
Comments: An iron-sulfur protein. Requires Mg2+ or Mn2+. Inactive towards aromatic acids that are not CoA esters but will also catalyse the reaction: ammonia + acceptor + 2 ADP + 2 phosphate = hydroxylamine + reduced acceptor + 2 ATP + H2O. In the presence of reduced acceptor, but in the absence of oxidizable substrate, the enzyme catalyses the hydrolysis of ATP to ADP plus phosphate.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, Metacyc, CAS registry number: 176591-18-7
References:
1. Boll, M. and Fuchs, G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234 (1995) 921-933. [PMID: 8575453]
2. Kung, J.W., Baumann, S., von Bergen, M., Muller, M., Hagedoorn, P.L., Hagen, W.R. and Boll, M. Reversible biological Birch reduction at an extremely low redox potential. J. Am. Chem. Soc. 132 (2010) 9850-9856. [PMID: 20578740]
[EC 1.3.7.9 Transferred entry: 4-hydroxybenzoyl-CoA reductase. Now classified as EC 1.1.7.1, 4-hydroxybenzoyl-CoA reductase. (EC 1.3.7.9 created 2000 as EC 1.3.99.20, transferred 2011 to EC 1.3.7.9, deleted 2020)]
[EC 1.3.7.10 Transferred entry: pentalenolactone synthase. Now classified as EC 1.14.19.8, pentalenolactone synthase (EC 1.3.7.10 created 2012, deleted 2013)]
Accepted name: 2,3-bis-O-geranylgeranyl-sn-glycero-phospholipid reductase
Reaction: a 2,3-bis-(O-phytanyl)-sn-glycero-phospholipid + 16 oxidized ferredoxin [iron-sulfur] cluster = a 2,3-bis-(O-geranylgeranyl)-sn-glycero-phospholipid + 16 reduced ferredoxin [iron-sulfur] cluster + 16 H+
For diagram of reaction click here.
Other name(s): AF0464 (gene name); 2,3-bis-O-geranylgeranyl-sn-glycerol 1-phosphate reductase (donor)
Systematic name: 2,3-bis-(O-phytanyl)-sn-glycero-phospholipid:ferredoxin oxidoreductase
Comments: A flavoprotein (FAD). The enzyme is involved in the biosynthesis of archaeal membrane lipids. It catalyses the reduction of all 8 double bonds in 2,3-bis-O-geranylgeranyl-sn-glycero-phospholipids and all 4 double bonds in 3-O-geranylgeranyl-sn-glycerol phospholipids with comparable activity. Unlike EC 1.3.1.101, 2,3-bis-O-geranylgeranyl-sn-glycerol 1-phosphate reductase [NAD(P)H], this enzyme shows no activity with NADPH, and requires a dedicated ferredoxin [4].
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Murakami, M., Shibuya, K., Nakayama, T., Nishino, T., Yoshimura, T. and Hemmi, H. Geranylgeranyl reductase involved in the biosynthesis of archaeal membrane lipids in the hyperthermophilic archaeon Archaeoglobus fulgidus. FEBS J. 274 (2007) 805-814. [PMID: 17288560]
2. Sato, S., Murakami, M., Yoshimura, T. and Hemmi, H. Specific partial reduction of geranylgeranyl diphosphate by an enzyme from the thermoacidophilic archaeon Sulfolobus acidocaldarius yields a reactive prenyl donor, not a dead-end product. J. Bacteriol. 190 (2008) 3923-3929. [PMID: 18375567]
3. Sasaki, D., Fujihashi, M., Iwata, Y., Murakami, M., Yoshimura, T., Hemmi, H. and Miki, K. Structure and mutation analysis of archaeal geranylgeranyl reductase. J. Mol. Biol. 409 (2011) 543-557. [PMID: 21515284]
4. Isobe, K., Ogawa, T., Hirose, K., Yokoi, T., Yoshimura, T. and Hemmi, H. Geranylgeranyl reductase and ferredoxin from Methanosarcina acetivorans are required for the synthesis of fully reduced archaeal membrane lipid in Escherichia coli cells. J. Bacteriol. 196 (2014) 417-423. [PMID: 24214941]
Accepted name: red chlorophyll catabolite reductase
Reaction: primary fluorescent chlorophyll catabolite + 2 oxidized ferredoxin [iron-sulfur] cluster = red chlorophyll catabolite + 2 reduced ferredoxin [iron-sulfur] cluster + 2 H+
For diagram of reaction click here.
Glossary: red chlorophyll catabolite = RCC = (7S,8S,101R)-8-(2-carboxyethyl)-17-ethyl-19-formyl-101-(methoxycarbonyl)-3,7,13,18-tetramethyl-2-vinyl-8,23-dihydro-7H-10,12-ethanobiladiene-ab-1,102(21H)-dione
primary fluorescent chlorophyll catabolite = pFCC = (82R,12S,13S)-12-(2-carboxyethyl)-3-ethyl-1-formyl-82-(methoxycarbonyl)-2,7,13,17-tetramethyl-18-vinyl-12,13-dihydro-8,10-ethanobilene-b-81,19(16H)-dione
Other name(s): RCCR; RCC reductase; red Chl catabolite reductase
Systematic name: primary fluorescent chlorophyll catabolite:ferredoxin oxidoreductase
Comments: The enzyme participates in chlorophyll degradation, which occurs during leaf senescence and fruit ripening in higher plants. The reaction requires reduced ferredoxin, which is generated from NADPH produced either through the pentose-phosphate pathway or by the action of photosystem I [1,2]. This reaction takes place while red chlorophyll catabolite is still bound to EC 1.14.15.17, pheophorbide a oxygenase [3]. Depending on the plant species used as the source of enzyme, one of two possible C-1 epimers of primary fluorescent chlorophyll catabolite (pFCC), pFCC-1 or pFCC-2, is normally formed, with all genera or species within a family producing the same isomer [3,4]. After modification and export, pFCCs are eventually imported into the vacuole, where the acidic environment causes their non-enzymic conversion into colourless breakdown products called non-fluorescent chlorophyll catabolites (NCCs) [2].
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Rodoni, S., Mühlecker, W., Anderl, M., Kräutler, B., Moser, D., Thomas, H., Matile, P. and Hörtensteiner, S. Chlorophyll breakdown in senescent chloroplasts. Cleavage of pheophorbide a in two enzymic steps. Plant Physiol. 115 (1997) 669-676. [PMID: 12223835]
2. Wüthrich, K.L., Bovet, L., Hunziker, P.E., Donnison, I.S. and Hörtensteiner, S. Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 21 (2000) 189-198. [PMID: 10743659]
3. Pružinská, A., Anders, I., Aubry, S., Schenk, N., Tapernoux-Lüthi, E., Müller, T., Kräutler, B. and Hörtensteiner, S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19 (2007) 369-387. [PMID: 17237353]
4. Hörtensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57 (2006) 55-77. [PMID: 16669755]
5. Rodoni, S., Vicentini, F., Schellenberg, M., Matile, P. and Hörtensteiner, S. Partial purification and characterization of red chlorophyll catabolite reductase, a stroma protein involved in chlorophyll breakdown. Plant Physiol. 115 (1997) 677-682. [PMID: 12223836]
Accepted name: 3,8-divinyl protochlorophyllide a 8-vinyl-reductase (ferredoxin)
Reaction: protochlorophyllide a + 2 oxidized ferredoxin [iron-sulfur] cluster = 3,8-divinyl protochlorophyllide a + 2 reduced ferredoxin [iron-sulfur] cluster + 2 H+
For diagram of reaction click here.
Other name(s): bciB (gene name); cyano-type divinyl chlorophyllide a 8-vinyl-reductase
Systematic name: protochlorophyllide-a:ferredoxin C-81-oxidoreductase
Comments: The enzyme, found in many phototrophic bacteria, land plants, and some green and red algae, is involved in the production of monovinyl versions of (bacterio)chlorophyll pigments from their divinyl precursors. Binds two [4Fe-4S] clusters and an FAD cofactor. It can also act on 3,8-divinyl chlorophyllide a, 3,8-divinyl chlorophyll a, and chlorophyll c2. cf. EC 1.3.1.75, 3,8-divinyl protochlorophyllide a 8-vinyl-reductase (NADPH).
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Chew, A.G. and Bryant, D.A. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J. Biol. Chem. 282 (2007) 2967-2975. [PMID: 17148453]
2. Saunders, A.H., Golbeck, J.H. and Bryant, D.A. Characterization of BciB: a ferredoxin-dependent 8-vinyl-protochlorophyllide reductase from the green sulfur bacterium Chloroherpeton thalassium. Biochemistry 52 (2013) 8442-8451. [PMID: 24151992]
3. Ito, H. and Tanaka, A. Evolution of a new chlorophyll metabolic pathway driven by the dynamic changes in enzyme promiscuous activity. Plant Cell Physiol 55 (2014) 593-603. [PMID: 24399236]
Accepted name: 3,8-divinyl chlorophyllide a reductase
Reaction: bacteriochlorophyllide g + 2 oxidized ferredoxin [iron-sulfur] cluster + ADP + phosphate = 3,8-divinyl chlorophyllide a + 2 reduced ferredoxin [iron-sulfur] cluster + ATP + H2O + 2 H+
For diagram of reaction click here.
Systematic name: bacteriochlorophyllide-g:ferredoxin C-81-oxidoreductase
Comments: The enzyme, found only in bacteriochlorophyll b-producing bacteria, catalyses the introduction of a C-8 ethylidene group. The enzyme contains a [4Fe-4S] cluster, and structurally resembles the Fe protein/MoFe protein complex of nitrogenase. It is very similar to EC 1.3.7.15, chlorophyllide a reductase, and is composed of three subunits. Two of them form the catalytic component, while the third one functions as an ATP-dependent reductase component that catalyses the electron transfer from ferredoxin to the catalytic component.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Tsukatani, Y., Yamamoto, H., Harada, J., Yoshitomi, T., Nomata, J., Kasahara, M., Mizoguchi, T., Fujita, Y. and Tamiaki, H. An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci Rep 3 (2013) 1217. [PMID: 23386973]
2. Tsukatani, Y., Harada, J., Nomata, J., Yamamoto, H., Fujita, Y., Mizoguchi, T. and Tamiaki, H. Rhodobacter sphaeroides mutants overexpressing chlorophyllide a oxidoreductase of Blastochloris viridis elucidate functions of enzymes in late bacteriochlorophyll biosynthetic pathways. Sci Rep 5 (2015) 9741. [PMID: 25978726]
Accepted name: chlorophyllide a reductase
Reaction: (1) 3-deacetyl-3-vinylbacteriochlorophyllide a + 2 oxidized ferredoxin [iron-sulfur] cluster + ADP + phosphate = chlorophyllide a + 2 reduced ferredoxin [iron-sulfur] cluster + ATP + H2O + 2 H+
(2) bacteriochlorophyllide a + 2 oxidized ferredoxin [iron-sulfur] cluster + ADP + phosphate = 3-acetyl-3-devinylchlorophyllide a + 2 reduced ferredoxin [iron-sulfur] cluster + ATP + H2O + 2 H+
(3) 3-deacetyl-3-(1-hydroxyethyl)bacteriochlorophyllide a + 2 oxidized ferredoxin [iron-sulfur] cluster + ADP + phosphate = 3-devinyl-3-(1-hydroxyethyl)chlorophyllide a + 2 reduced ferredoxin [iron-sulfur] cluster + ATP + H2O + 2 H+
For diagram of reaction click here.
Other name(s): bchX (gene name); bchY (gene name); bchZ (gene name); COR
Systematic name: bacteriochlorophyllide-a:ferredoxin 7,8-oxidoreductase
Comments: The enzyme, together with EC 1.1.1.396, bacteriochlorophyllide-a dehydrogenase, and EC 4.2.1.165, chlorophyllide-a 31-hydratase, is involved in the conversion of chlorophyllide a to bacteriochlorophyllide a. These enzymes can act in multiple orders, resulting in the formation of different intermediates, but the final product of the cumulative action of the three enzymes is always bacteriochlorophyllide a. This enzyme catalyses a trans-reduction of the B-ring; the product has the (7R,8R)-configuration. In addition, the enzyme has a latent activity of EC 1.3.7.13, 3,8-divinyl protochlorophyllide a 8-vinyl-reductase (ferredoxin) [4]. The enzyme contains a [4Fe-4S] cluster, and structurally resembles the Fe protein/MoFe protein complex of nitrogenase (EC 1.18.6.1), which catalyses an ATP-driven reduction.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Nomata, J., Mizoguchi, T., Tamiaki, H. and Fujita, Y. A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis: reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281 (2006) 15021-15028. [PMID: 16571720]
2. Tsukatani, Y., Yamamoto, H., Harada, J., Yoshitomi, T., Nomata, J., Kasahara, M., Mizoguchi, T., Fujita, Y. and Tamiaki, H. An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci Rep 3 (2013) 1217. [PMID: 23386973]
3. Lange, C., Kiesel, S., Peters, S., Virus, S., Scheer, H., Jahn, D. and Moser, J. Broadened substrate specificity of 3-hydroxyethyl bacteriochlorophyllide a dehydrogenase (BchC) indicates a new route for the biosynthesis of bacteriochlorophyll a. J. Biol. Chem. 290 (2015) 19697-19709. [PMID: 26088139]
4. Harada, J., Mizoguchi, T., Tsukatani, Y., Yokono, M., Tanaka, A. and Tamiaki, H. Chlorophyllide a oxidoreductase works as one of the divinyl reductases specifically involved in bacteriochlorophyll a biosynthesis. J. Biol. Chem. 289 (2014) 12716-12726. [PMID: 24637023]
Accepted name: short-chain acyl-CoA dehydrogenase
Reaction: a short-chain acyl-CoA + electron-transfer flavoprotein = a short-chain trans-2,3-dehydroacyl-CoA + reduced electron-transfer flavoprotein
Glossary: a short-chain acyl-CoA = an acyl-CoA thioester where the acyl chain contains less than 6 carbon atoms.
Other name(s): butyryl-CoA dehydrogenase; butanoyl-CoA dehydrogenase; butyryl dehydrogenase; unsaturated acyl-CoA reductase; ethylene reductase; enoyl-coenzyme A reductase; unsaturated acyl coenzyme A reductase; butyryl coenzyme A dehydrogenase; short-chain acyl CoA dehydrogenase; short-chain acyl-coenzyme A dehydrogenase; 3-hydroxyacyl CoA reductase; butanoyl-CoA:(acceptor) 2,3-oxidoreductase; ACADS (gene name).
Systematic name: short-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: Contains a tightly-bound FAD cofactor. One of several enzymes that catalyse the first step in fatty acids β-oxidation. The enzyme catalyses the oxidation of saturated short-chain acyl-CoA thioesters to give a trans 2,3-unsaturated product by removal of the two pro-R-hydrogen atoms. The enzyme from beef liver accepts substrates with acyl chain lengths of 3 to 8 carbon atoms. The highest activity was reported with either butanoyl-CoA [2] or pentanoyl-CoA [4]. The enzyme from rat has only 10% activity with hexanoyl-CoA (compared to butanoyl-CoA) and no activity with octanoyl-CoA [6]. cf. EC 1.3.8.7, medium-chain acyl-CoA dehydrogenase, EC 1.3.8.8, long-chain acyl-CoA dehydrogenase, and EC 1.3.8.9, very-long-chain acyl-CoA dehydrogenase.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, GTD, KEGG, Metacyc, PDB, CAS registry number: 9027-88-7
References:
1. Mahler, H.R. Studies on the fatty acid oxidizing system of animal tissue. IV. The prosthetic group of butyryl coenzyme A dehydrogenase. J. Biol. Chem. 206 (1954) 13-26. [PMID: 13130522]
2. Green, D.E., Mii, S., Mahler, H.R. and Bock, R.M. Studies on the fatty acid oxidizing system of animal tissue. III. Butyryl coenzyme A dehydrogenase. J. Biol. Chem. 206 (1954) 1-12. [PMID: 13130521]
3. Beinert, H. Acyl coenzyme A dehydrogenase. In: Boyer, P.D., Lardy, H. and Myrbäck, K. (Eds), The Enzymes, 2nd edn, vol. 7, Academic Press, New York, 1963, pp. 447-466.
4. Shaw, L. and Engel, P.C. The purification and properties of ox liver short-chain acyl-CoA dehydrogenase. Biochem. J. 218 (1984) 511-520. [PMID: 6712627]
5. Thorpe, C. and Kim, J.J. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 9 (1995) 718-725. [PMID: 7601336]
6. Ikeda, Y., Ikeda, K.O. and Tanaka, K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J. Biol. Chem. 260 (1985) 1311-1325. [PMID: 3968063]
7. McMahon, B., Gallagher, M.E. and Mayhew, S.G. The protein coded by the PP2216 gene of Pseudomonas putida KT2440 is an acyl-CoA dehydrogenase that oxidises only short-chain aliphatic substrates. FEMS Microbiol. Lett. 250 (2005) 121-127. [PMID: 16024185]
Accepted name: 4,4'-diapophytoene desaturase
Reaction: 15-cis-4,4'-diapophytoene + 4 FAD = all-trans-4,4'-diapolycopene + 4 FADH2 (overall reaction)
(1a) 15-cis-4,4'-diapophytoene + FAD = all-trans-4,4'-diapophytofluene + FADH2
(1b) all-trans-4,4'-diapophytofluene + FAD = all-trans-4,4'-diapo-ζ-carotene + FADH2
(1c) all-trans-4,4'-diapo-ζ-carotene + FAD = all-trans-4,4'-diaponeurosporene + FADH2
(1d) all-trans-4,4'-diaponeurosporene + FAD = all-trans-4,4'-diapolycopene + FADH2
For diagram of reaction click here
Other name(s): dehydrosqualene desaturase; CrtN; 4,4'-diapophytoene:FAD oxidoreductase
Systematic name: 15-cis-4,4'-diapophytoene:FAD oxidoreductase
Comments: Typical of Staphylococcus aureus and some other bacteria such as Heliobacillus sp. Responsible for four successive dehydrogenations. In some species it only proceeds as far as all-trans-4,4'-diaponeurosporene.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Wieland, B., Feil, C., Gloria-Maercker, E., Thumm, G., Lechner, M., Bravo, J.M., Poralla, K. and Gotz, F. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4'-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176 (1994) 7719-7726. [PMID: 8002598]
2. Raisig, A. and Sandmann, G. 4,4'-diapophytoene desaturase: catalytic properties of an enzyme from the C30 carotenoid pathway of Staphylococcus aureus. J. Bacteriol. 181 (1999) 6184-6187. [PMID: 10498735]
3. Raisig, A. and Sandmann, G. Functional properties of diapophytoene and related desaturases of C30 to C40 carotenoid biosynthetic pathways. Biochim. Biophys. Acta 1533 (2001) 164-170. [PMID: 11566453]
Accepted name: (R)-benzylsuccinyl-CoA dehydrogenase
Reaction: (R)-2-benzylsuccinyl-CoA + electron-transfer flavoprotein = (E)-2-benzylidenesuccinyl-CoA + reduced electron-transfer flavoprotein
For diagram of reaction, click here
Other name(s): BbsG; (R)-benzylsuccinyl-CoA:(acceptor) oxidoreductase
Systematic name: (R)-benzylsuccinyl-CoA:electron transfer flavoprotein oxidoreductase
Comments: Requires FAD as prosthetic group. Unlike other acyl-CoA dehydrogenases, this enzyme exhibits high substrate- and enantiomer specificity; it is highly specific for (R)-benzylsuccinyl-CoA and is inhibited by (S)-benzylsuccinyl-CoA. Forms the third step in the anaerobic toluene metabolic pathway in Thauera aromatica. Ferricenium ion is an effective artificial electron acceptor.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Leutwein, C. and Heider, J. Anaerobic toluene-catabolic pathway in denitrifying Thauera aromatica: activation and β-oxidation of the first intermediate, (R)-(+)-benzylsuccinate. Microbiology 145 (1999) 3265-3271. [PMID: 10589736]
2. Leutwein, C. and Heider, J. (R)-Benzylsuccinyl-CoA dehydrogenase of Thauera aromatica, an enzyme of the anaerobic toluene catabolic pathway. Arch. Microbiol. 178 (2002) 517-524. [PMID: 12420174]
Accepted name: isovaleryl-CoA dehydrogenase
Reaction: isovaleryl-CoA + electron-transfer flavoprotein = 3-methylcrotonyl-CoA + reduced electron-transfer flavoprotein
Other name(s): isovaleryl-coenzyme A dehydrogenase; isovaleroyl-coenzyme A dehydrogenase; 3-methylbutanoyl-CoA:(acceptor) oxidoreductase
Systematic name: 3-methylbutanoyl-CoA:electron-transfer flavoprotein oxidoreductase
Comments: Contains FAD as prosthetic group. Pentanoate can act as donor.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 37274-61-6
References:
1. Bachhawat, B.K., Robinson, W.G. and Coon, M.J. Enzymatic carboxylation of β-hydroxyisovaleryl coenzyme A. J. Biol. Chem. 219 (1956) 539-550. [PMID: 13319276]
2. Ikeda, Y. and Tanaka, K. Purification and characterization of isovaleryl coenzyme A dehydrogenase from rat liver mitochondria. J. Biol. Chem. 258 (1983) 1077-1085. [PMID: 6401713]
3. Tanaka, K., Budd, M.A., Efron, M.L. and Isselbacher, K.J. Isovaleric acidemia: a new genetic defect of leucine metabolism. Proc. Natl. Acad. Sci. USA 56 (1966) 236-242. [PMID: 5229850]
Accepted name: short-chain 2-methylacyl-CoA dehydrogenase
Reaction: 2-methylbutanoyl-CoA + electron-transfer flavoprotein = (E)-2-methylbut-2-enoyl-CoA + reduced electron-transfer flavoprotein + H+
Other name(s): ACADSB (gene name); 2-methylacyl-CoA dehydrogenase; branched-chain acyl-CoA dehydrogenase (ambiguous); 2-methyl branched chain acyl-CoA dehydrogenase; 2-methylbutanoyl-CoA:(acceptor) oxidoreductase; 2-methyl-branched-chain-acyl-CoA:electron-transfer flavoprotein 2-oxidoreductase; 2-methyl-branched-chain-enoyl-CoA reductase
Systematic name: short-chain 2-methylacyl-CoA:electron-transfer flavoprotein 2-oxidoreductase
Comments: A flavoprotein (FAD). The mammalian enzyme catalyses an oxidative reaction as a step in the mitochondrial β-oxidation of short-chain 2-methyl fatty acids and participates in isoleucine degradation. The enzyme from the parasitic helminth Ascaris suum catalyses a reductive reaction as part of a fermentation pathway, shuttling reducing power from the electron-transport chain to 2-methyl branched-chain enoyl CoA.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Ikeda, Y., Dabrowski, C. and Tanaka, K. Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification of a new 2-methyl branched chain acyl-CoA dehydrogenase. J. Biol. Chem. 258 (1983) 1066-1076. [PMID: 6401712]
2. Komuniecki, R., Fekete, S. and Thissen-Parra, J. Purification and characterization of the 2-methyl branched-chain acyl-CoA dehydrogenase, an enzyme involved in NADH-dependent enoyl-CoA reduction in anaerobic mitochondria of the nematode, Ascaris suum. J. Biol. Chem. 260 (1985) 4770-4777. [PMID: 3988734]
3. Komuniecki, R., McCrury, J., Thissen, J. and Rubin, N. Electron-transfer flavoprotein from anaerobic Ascaris suum mitochondria and its role in NADH-dependent 2-methyl branched-chain enoyl-CoA reduction. Biochim. Biophys. Acta 975 (1989) 127-131. [PMID: 2736251]
4. Vockley, J., Mohsen al,-W., A., Binzak, B., Willard, J. and Fauq, A. Mammalian branched-chain acyl-CoA dehydrogenases: molecular cloning and characterization of recombinant enzymes. Methods Enzymol. 324 (2000) 241-258. [PMID: 10989435]
5. Andresen, B.S., Christensen, E., Corydon, T.J., Bross, P., Pilgaard, B., Wanders, R.J., Ruiter, J.P., Simonsen, H., Winter, V., Knudsen, I., Schroeder, L.D., Gregersen, N. and Skovby, F. Isolated 2-methylbutyrylglycinuria caused by short/branched-chain acyl-CoA dehydrogenase deficiency: identification of a new enzyme defect, resolution of its molecular basis, and evidence for distinct acyl-CoA dehydrogenases in isoleucine and valine metabolism. Am. J. Hum. Genet. 67 (2000) 1095-1103. [PMID: 11013134]
Accepted name: glutaryl-CoA dehydrogenase (ETF)
Reaction: glutaryl-CoA + electron-transfer flavoprotein = crotonyl-CoA + CO2 + reduced electron-transfer flavoprotein (overall reaction)
(1a) glutaryl-CoA + electron-transfer flavoprotein = (E)-glutaconyl-CoA + reduced electron-transfer flavoprotein
(1b) (E)-glutaconyl-CoA = crotonyl-CoA + CO2
For diagram of reaction click here.
Glossary: (E)-glutaconyl-CoA = (2E)-4-carboxybut-2-enoyl-CoA
crotonyl-CoA = (E)-but-2-enoyl-CoA
Other name(s): glutaryl coenzyme A dehydrogenase; glutaryl-CoA:(acceptor) 2,3-oxidoreductase (decarboxylating); glutaryl-CoA dehydrogenase
Systematic name: glutaryl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase (decarboxylating)
Comments: Contains FAD. The enzyme catalyses the oxidation of glutaryl-CoA to glutaconyl-CoA (which remains bound to the enzyme), and the decarboxylation of the latter to crotonyl-CoA (cf. EC 7.2.4.5, glutaconyl-CoA decarboxylase). FAD is the electron acceptor in the oxidation of the substrate, and its reoxidation by electron-transfer flavoprotein completes the catalytic cycle. The anaerobic, sulfate-reducing bacterium Desulfococcus multivorans contains two glutaryl-CoA dehydrogenases: a decarboxylating enzyme (this entry), and a non-decarboxylating enzyme that only catalyses the oxidation to glutaconyl-CoA [EC 1.3.99.32, glutaryl-CoA dehydrogenase (acceptor)].
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, ExplorEnz, KEGG, MetaCyc, PDB, CAS registry number: 37255-38-2
References:
1. Besrat, A., Polan, C.E. and Henderson, L.M. Mammalian metabolism of glutaric acid. J. Biol. Chem. 244 (1969) 1461-1467. [PMID: 4304226]
2. Hartel, U., Eckel, E., Koch, J., Fuchs, G., Linder, D. and Buckel, W. Purification of glutaryl-CoA dehydrogenase from Pseudomonas sp., an enzyme involved in the anaerobic degradation of benzoate. Arch. Microbiol. 159 (1993) 174-181. [PMID: 8439237]
3. Dwyer, T.M., Zhang, L., Muller, M., Marrugo, F. and Frerman, F. The functions of the flavin contact residues, αArg249 and βTyr16, in human electron transfer flavoprotein. Biochim. Biophys. Acta 1433 (1999) 139-152. [PMID: 10446367]
4. Rao, K.S., Albro, M., Dwyer, T.M. and Frerman, F.E. Kinetic mechanism of glutaryl-CoA dehydrogenase. Biochemistry 45 (2006) 15853-15861. [PMID: 17176108]
Accepted name: medium-chain acyl-CoA dehydrogenase
Reaction: a medium-chain acyl-CoA + electron-transfer flavoprotein = a medium-chain trans-2,3-dehydroacyl-CoA + reduced electron-transfer flavoprotein
Glossary: a medium-chain acyl-CoA = an acyl-CoA thioester where the acyl chain contains 6 to 12 carbon atoms.
Other name(s): fatty acyl coenzyme A dehydrogenase (ambiguous); acyl coenzyme A dehydrogenase (ambiguous); acyl dehydrogenase (ambiguous); fatty-acyl-CoA dehydrogenase (ambiguous); acyl CoA dehydrogenase (ambiguous); general acyl CoA dehydrogenase (ambiguous); medium-chain acyl-coenzyme A dehydrogenase; acyl-CoA:(acceptor) 2,3-oxidoreductase (ambiguous); ACADM (gene name).
Systematic name: medium-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: Contains FAD as prosthetic group. One of several enzymes that catalyse the first step in fatty acids β-oxidation. The enzyme from pig liver can accept substrates with acyl chain lengths of 4 to 16 carbon atoms, but is most active with C8 to C12 compounds [2]. The enzyme from rat does not accept C16 at all and is most active with C6-C8 compounds [4]. cf. EC 1.3.8.1, short-chain acyl-CoA dehydrogenase, EC 1.3.8.8, long-chain acyl-CoA dehydrogenase, and EC 1.3.8.9, very-long-chain acyl-CoA dehydrogenase.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Crane, F.L., Hauge, J.G. and Beinert, H. Flavoproteins involved in the first oxidative step of the fatty acid cycle. Biochim. Biophys. Acta 17 (1955) 292-294. [PMID: 13239683]
2. Crane, F.L., Mii, S., Hauge, J.G., Green, D.E. and Beinert, H. On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. I. The general fatty acyl coenzyme A dehydrogenase. J. Biol. Chem. 218 (1956) 701-716. [PMID: 13295224]
3. Beinert, H. Acyl coenzyme A dehydrogenase. In: Boyer, P.D., Lardy, H. and Myrbäck, K. (Eds), The Enzymes, 2nd edn, vol. 7, Academic Press, New York, 1963, pp. 447-466.
4. Ikeda, Y., Ikeda, K.O. and Tanaka, K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J. Biol. Chem. 260 (1985) 1311-1325. [PMID: 3968063]
5. Thorpe, C. and Kim, J.J. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 9 (1995) 718-725. [PMID: 7601336]
6. Kim, J.J., Wang, M. and Paschke, R. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc. Natl. Acad. Sci. USA 90 (1993) 7523-7527. [PMID: 8356049]
7. Peterson, K.L., Sergienko, E.E., Wu, Y., Kumar, N.R., Strauss, A.W., Oleson, A.E., Muhonen, W.W., Shabb, J.B. and Srivastava, D.K. Recombinant human liver medium-chain acyl-CoA dehydrogenase: purification, characterization, and the mechanism of interactions with functionally diverse C8-CoA molecules. Biochemistry 34 (1995) 14942-14953. [PMID: 7578106]
8. Toogood, H.S., van Thiel, A., Basran, J., Sutcliffe, M.J., Scrutton, N.S. and Leys, D. Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex. J. Biol. Chem. 279 (2004) 32904-32912. [PMID: 15159392]
Accepted name: long-chain acyl-CoA dehydrogenase
Reaction: a long-chain acyl-CoA + electron-transfer flavoprotein = a long-chain trans-2,3-dehydroacyl-CoA + reduced electron-transfer flavoprotein
Glossary: a long-chain acyl-CoA = an acyl-CoA thioester where the acyl chain contains 13 to 22 carbon atoms.
Other name(s): palmitoyl-CoA dehydrogenase; palmitoyl-coenzyme A dehydrogenase; long-chain acyl-coenzyme A dehydrogenase; long-chain-acyl-CoA:(acceptor) 2,3-oxidoreductase; ACADL (gene name).
Systematic name: long-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: Contains FAD as prosthetic group. One of several enzymes that catalyse the first step in fatty acids β-oxidation. The enzyme from pig liver can accept substrates with acyl chain lengths of 6 to at least 16 carbon atoms. The highest activity was found with C12, and the rates with C8 and C16 were 80 and 70%, respectively [2]. The enzyme from rat can accept substrates with C8-C22. It is most active with C14 and C16, and has no activity with C4, C6 or C24 [4]. cf. EC 1.3.8.1, short-chain acyl-CoA dehydrogenase, EC 1.3.8.8, medium-chain acyl-CoA dehydrogenase, and EC 1.3.8.9, very-long-chain acyl-CoA dehydrogenase.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 59536-74-2
References:
1. Crane, F.L., Hauge, J.G. and Beinert, H. Flavoproteins involved in the first oxidative step of the fatty acid cycle. Biochim. Biophys. Acta 17 (1955) 292-294. [PMID: 13239683]
2. Hauge, J.G., Crane, F.L. and Beinert, H. On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. III. Palmityl CoA dehydrogenase. J. Biol. Chem. 219 (1956) 727-733. [PMID: 13319294]
3. Hall, C.L., Heijkenkjold, L., Bartfai, T., Ernster, L. and Kamin, H. Acyl coenzyme A dehydrogenases and electron-transferring flavoprotein from beef heart mitochondria. Arch. Biochem. Biophys. 177 (1976) 402-414. [PMID: 1015826]
4. Ikeda, Y., Ikeda, K.O. and Tanaka, K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J. Biol. Chem. 260 (1985) 1311-1325. [PMID: 3968063]
5. Djordjevic, S., Dong, Y., Paschke, R., Frerman, F.E., Strauss, A.W. and Kim, J.J. Identification of the catalytic base in long chain acyl-CoA dehydrogenase. Biochemistry 33 (1994) 4258-4264. [PMID: 8155643]
Accepted name: very-long-chain acyl-CoA dehydrogenase
Reaction: a very-long-chain acyl-CoA + electron-transfer flavoprotein = a very-long-chain trans-2,3-dehydroacyl-CoA + reduced electron-transfer flavoprotein
Glossary: a very-long-chain acyl-CoA = an acyl-CoA thioester where the acyl chain contains 23 or more carbon atoms.
Other name(s): ACADVL (gene name).
Systematic name: very-long-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: Contains FAD as prosthetic group. One of several enzymes that catalyse the first step in fatty acids β-oxidation. The enzyme is most active toward long-chain acyl-CoAs such as C14, C16 and C18, but is also active with very-long-chain acyl-CoAs up to 24 carbons. It shows no activity for substrates of less than 12 carbons. Its specific activity towards palmitoyl-CoA is more than 10-fold that of the long-chain acyl-CoA dehydrogenase [1]. cf. EC 1.3.8.1, short-chain acyl-CoA dehydrogenase, EC 1.3.8.7, medium-chain acyl-CoA dehydrogenase, and EC 1.3.8.8, long-chain acyl-CoA dehydrogenase.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Izai, K., Uchida, Y., Orii, T., Yamamoto, S. and Hashimoto, T. Novel fatty acid β-oxidation enzymes in rat liver mitochondria. I. Purification and properties of very-long-chain acyl-coenzyme A dehydrogenase. J. Biol. Chem. 267 (1992) 1027-1033. [PMID: 1730632]
2. Aoyama, T., Souri, M., Ushikubo, S., Kamijo, T., Yamaguchi, S., Kelley, R.I., Rhead, W.J., Uetake, K., Tanaka, K. and Hashimoto, T. Purification of human very-long-chain acyl-coenzyme A dehydrogenase and characterization of its deficiency in seven patients. J. Clin. Invest. 95 (1995) 2465-2473. [PMID: 7769092]
3. McAndrew, R.P., Wang, Y., Mohsen, A.W., He, M., Vockley, J. and Kim, J.J. Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase. J. Biol. Chem. 283 (2008) 9435-9443. [PMID: 18227065]
Accepted name: cyclohex-1-ene-1-carbonyl-CoA dehydrogenase
Reaction: cyclohex-1-ene-1-carbonyl-CoA + electron-transfer flavoprotein = cyclohex-1,5-diene-1-carbonyl-CoA + reduced electron-transfer flavoprotein
Systematic name: cyclohex-1-ene-1-carbonyl-CoA:electron transfer flavoprotein oxidoreductase
Comments: Contains FAD. The enzyme, characterized from the strict anaerobic bacterium Syntrophus aciditrophicus, is involved in production of cyclohexane-1-carboxylate, a byproduct produced by that organism during fermentation of benzoate and crotonate to acetate.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Kung, J.W., Seifert, J., von Bergen, M. and Boll, M. Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in the fermentation of benzoate and crotonate in Syntrophus aciditrophicus. J. Bacteriol. 195 (2013) 3193-3200. [PMID: 23667239]
Accepted name: cyclohexane-1-carbonyl-CoA dehydrogenase (electron-transfer flavoprotein)
Reaction: cyclohexane-1-carbonyl-CoA + electron-transfer flavoprotein = cyclohex-1-ene-1-carbonyl-CoA + reduced electron-transfer flavoprotein
Other name(s): aliB (gene name); cyclohexane-1-carbonyl-CoA dehydrogenase (ambiguous)
Systematic name: cyclohexane-1-carbonyl-CoA:electron transfer flavoprotein oxidoreductase
Comments: Contains FAD. The enzyme, characterized from the strict anaerobic bacterium Syntrophus aciditrophicus, is involved in production of cyclohexane-1-carboxylate, a byproduct produced by that organism during fermentation of benzoate and crotonate to acetate.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Pelletier, D.A. and Harwood, C.S. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J. Bacteriol. 182 (2000) 2753-2760. [PMID: 10781543]
2. Kung, J.W., Seifert, J., von Bergen, M. and Boll, M. Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in the fermentation of benzoate and crotonate in Syntrophus aciditrophicus. J. Bacteriol. 195 (2013) 3193-3200. [PMID: 23667239]
Accepted name: (2S)-methylsuccinyl-CoA dehydrogenase
Reaction: (2S)-methylsuccinyl-CoA + electron-transfer flavoprotein = 2-methylfumaryl-CoA + reduced electron-transfer flavoprotein
Glossary: 2-methylfumaryl-CoA = (E)-3-carboxy-2-methylprop-2-enoyl-CoA
Other name(s): Mcd
Systematic name: (2S)-methylsuccinyl-CoA:electron-transfer flavoprotein oxidoreductase
Comments: The enzyme, characterized from the bacterium Rhodobacter sphaeroides, is involved in the ethylmalonyl-CoA pathway for acetyl-CoA assimilation. The enzyme contains FAD.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Erb, T.J., Fuchs, G. and Alber, B.E. (2S)-Methylsuccinyl-CoA dehydrogenase closes the ethylmalonyl-CoA pathway for acetyl-CoA assimilation. Mol. Microbiol. 73 (2009) 992-1008. [PMID: 19703103]
Accepted name: crotonobetainyl-CoA reductase
Reaction: γ-butyrobetainyl-CoA + electron-transfer flavoprotein = crotonobetainyl-CoA + reduced electron-transfer flavoprotein
Glossary: γ-butyrobetainyl-CoA = 4-(trimethylammonio)butanoyl-CoA
crotonobetainyl-CoA = (E)-4-(trimethylammonio)but-2-enoyl-CoA
Other name(s): caiA (gene name)
Systematic name: γ-butyrobetainyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: The enzyme has been purified from the bacterium Escherichia coli O44 K74, in which it forms a complex with EC 2.8.3.21, L-carnitine CoA-transferase. The electron donor is believed to be an electron-transfer flavoprotein (ETF) encoded by the fixA and fixB genes.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Roth, S., Jung, K., Jung, H., Hommel, R.K. and Kleber, H.P. Crotonobetaine reductase from Escherichia coli - a new inducible enzyme of anaerobic metabolization of L(–)-carnitine. Antonie Van Leeuwenhoek 65 (1994) 63-69. [PMID: 8060125]
2. Preusser, A., Wagner, U., Elssner, T. and Kleber, H.P. Crotonobetaine reductase from Escherichia coli consists of two proteins. Biochim. Biophys. Acta 1431 (1999) 166-178. [PMID: 10209289]
3. Elssner, T., Hennig, L., Frauendorf, H., Haferburg, D. and Kleber, H.P. Isolation, identification, and synthesis of γ-butyrobetainyl-CoA and crotonobetainyl-CoA, compounds involved in carnitine metabolism of E. coli. Biochemistry 39 (2000) 10761-10769. [PMID: 10978161]
4. Walt, A. and Kahn, M.L. The fixA and fixB genes are necessary for anaerobic carnitine reduction in Escherichia coli. J. Bacteriol. 184 (2002) 4044-4047. [PMID: 12081978]
Accepted name: L-prolyl-S-[peptidyl-carrier protein] dehydrogenase
Reaction: S-(L-prolyl)-[peptidyl-carrier protein] + 2 electron-transfer flavoprotein = S-(1H-pyrrole-2-carbonyl)-[peptidyl-carrier protein] + 2 reduced electron-transfer flavoprotein
Other name(s): pigA (gene name); bmp3 (gene name); pltE (gene name); redW (gene name)
Systematic name: S-(L-prolyl)-[peptidyl-carrier protein]:electron-transfer flavoprotein oxidoreductase
Comments: Contains FAD. The enzyme participates in the biosynthesis of several pyrrole-containing compounds, such as undecylprodigiosin, prodigiosin, pyoluteorin, and coumermycin A1. It is believed to catalyse the formation of a Δ2-pyrrolin-2-yl(carbonyl)-S-[peptidyl-carrier protein] intermediate, followed by a two-electron oxidation to 1H-pyrrol-2-yl(carbonyl)-S-[peptidyl-carrier protein].
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Thomas, M.G., Burkart, M.D. and Walsh, C.T. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 9 (2002) 171-184. [PMID: 11880032]
2. Harris, A.K., Williamson, N.R., Slater, H., Cox, A., Abbasi, S., Foulds, I., Simonsen, H.T., Leeper, F.J. and Salmond, G.P. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150 (2004) 3547-3560. [PMID: 15528645]
Accepted name: 3-(aryl)acrylate reductase
Reaction: (1) phloretate + electron-transfer flavoprotein = 4-coumarate + reduced electron-transfer flavoprotein
(2) 3-phenylpropanoate + electron-transfer flavoprotein = trans-cinnamate + reduced electron-transfer flavoprotein
(3) 3-(1H-indol-3-yl)propanoate + electron-transfer flavoprotein = 3-(indol-3-yl)acrylate + reduced electron-transfer flavoprotein
Glossary: phloretate = 3-(4-hydroxyphenyl)propanoate
crotonate = (2E)-but-2-enoate
Other name(s): acdA (gene name)
Systematic name: 3-(phenyl)propanoate:electron-transfer flavoprotein 2,3-oxidoreductase
Comments: The enzyme, found in some amino acid-fermenting anaerobic bacteria, participates in the fermentation pathways of L-phenylalanine, L-tyrosine, and L-tryptophan. Unlike EC 1.3.1.31, 2-enoate reductase, this enzyme has minimal activity with crotonate.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Dodd, D., Spitzer, M.H., Van Treuren, W., Merrill, B.D., Hryckowian, A.J., Higginbottom, S.K., Le, A., Cowan, T.M., Nolan, G.P., Fischbach, M.A. and Sonnenburg, J.L. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551 (2017) 648-652. [PMID: 29168502]
Accepted name: 2-amino-4-deoxychorismate dehydrogenase
Reaction: (2S)-2-amino-4-deoxychorismate + FMN = 3-(1-carboxyvinyloxy)anthranilate + FMNH2
For diagram of reaction click here
Glossary: (2S)-2-amino-4-deoxychorismate = (2S,3S)-3-(1-carboxyvinyloxy)-2,3-dihydroanthranilate
3-enolpyruvoylanthranilate = 3-(1-carboxyvinyloxy)anthranilate
Other name(s): ADIC dehydrogenase; 2-amino-2-deoxyisochorismate dehydrogenase; SgcG
Systematic name: (2S)-2-amino-4-deoxychorismate:FMN oxidoreductase
Comments: The sequential action of EC 2.6.1.86, 2-amino-4-deoxychorismate synthase and this enzyme leads to the formation of the benzoxazolinate moiety of the enediyne antitumour antibiotic C-1027 [1,2].
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Van Lanen, S.G., Lin, S. and Shen, B. Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. Proc. Natl. Acad. Sci. USA 105 (2008) 494-499. [PMID: 18182490]
2. Yu, L., Mah, S., Otani, T. and Dedon, P. The benzoxazolinate of C-1027 confers intercalative DNA binding. J. Am. Chem. Soc. 117 (1995) 8877-8878.
Accepted name: dehydro coenzyme F420 reductase
Reaction: oxidized coenzyme F420-0 + FMN = dehydro coenzyme F420-0 + FMNH2
Glossary: dehydro coenzyme F420-0 = 2-{[5-deoxy-5-(8-hydroxy-2,4-dioxopyrimidino[4,5-b]quinolin-10(2H)-yl)-L-ribityloxy]hydroxyphosphoryloxy}prop-2-enoate
Other name(s): fbiB (gene name)
Systematic name: oxidized coenzyme F420-0:FMN oxidoreductase
Comments: This enzyme is involved in the biosynthesis of coenzyme F420, a redox-active cofactor found in all methanogenic archaea, as well as some eubacteria. In some eubacteria the enzyme is multifunctional, also catalysing the activities of EC 6.3.2.31, coenzyme F420-0:L-glutamate ligase, and EC 6.3.2.34, coenzyme F420-1:γ-L-glutamate ligase.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Bashiri, G., Antoney, J., Jirgis, E.NM., Shah, M.V., Ney, B., Copp, J., Stuteley, S.M., Sreebhavan, S., Palmer, B., Middleditch, M., Tokuriki, N., Greening, C., Scott, C., Baker, E.N. and Jackson, C.J. A revised biosynthetic pathway for the cofactor F420 in prokaryotes. Nat. Commun. 10 (2019) 1558. [PMID: 30952857]
Accepted name: dihydroorotate dehydrogenase (fumarate)
Reaction: (S)-dihydroorotate + fumarate = orotate + succinate
Other name(s): DHOdehase (ambiguous); dihydroorotate dehydrogenase (ambiguous); dihydoorotic acid dehydrogenase (ambiguous); DHOD (ambiguous); DHODase (ambiguous); dihydroorotate oxidase; pyr4 (gene name)
Systematic name: (S)-dihydroorotate:fumarate oxidoreductase
Comments: Binds FMN. The reaction, which takes place in the cytosol, is the only redox reaction in the de novo biosynthesis of pyrimidine nucleotides. Molecular oxygen can replace fumarate in vitro. Other class 1 dihydroorotate dehydrogenases use either NAD+ (EC 1.3.1.14) or NADP+ (EC 1.3.1.15) as electron acceptor. The membrane bound class 2 dihydroorotate dehydrogenase (EC 1.3.5.2) uses quinone as electron acceptor.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9029-03-2
References:
1. Björnberg, O., Rowland, P., Larsen, S. and Jensen, K.F. Active site of dihydroorotate dehydrogenase A from Lactococcus lactis investigated by chemical modification and mutagenesis. Biochemistry 36 (1997) 16197-16205. [PMID: 9405053]
2. Rowland, P., Björnberg, O., Nielsen, F.S., Jensen, K.F. and Larsen, S. The crystal structure of Lactococcus lactis dihydroorotate dehydrogenase A complexed with the enzyme reaction product throws light on its enzymatic function. Protein Sci. 7 (1998) 1269-1279. [PMID: 9655329]
3. Nørager, S., Arent, S., Björnberg, O., Ottosen, M., Lo Leggio, L., Jensen, K.F. and Larsen, S. Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function. J. Biol. Chem. 278 (2003) 28812-28822. [PMID: 12732650]
4. Zameitat, E., Pierik, A.J., Zocher, K. and Löffler, M. Dihydroorotate dehydrogenase from Saccharomyces cerevisiae: spectroscopic investigations with the recombinant enzyme throw light on catalytic properties and metabolism of fumarate analogues. FEMS Yeast Res. 7 (2007) 897-904. [PMID: 17617217]
5. Inaoka, D.K., Sakamoto, K., Shimizu, H., Shiba, T., Kurisu, G., Nara, T., Aoki, T., Kita, K. and Harada, S. Structures of Trypanosoma cruzi dihydroorotate dehydrogenase complexed with substrates and products: atomic resolution insights into mechanisms of dihydroorotate oxidation and fumarate reduction. Biochemistry 47 (2008) 10881-10891. [PMID: 18808149]
6. Cheleski, J., Wiggers, H.J., Citadini, A.P., da Costa Filho, A.J., Nonato, M.C. and Montanari, C.A. Kinetic mechanism and catalysis of Trypanosoma cruzi dihydroorotate dehydrogenase enzyme evaluated by isothermal titration calorimetry. Anal. Biochem. 399 (2010) 13-22. [PMID: 19932077]
[EC 1.3.98.2 Transferred entry: fumarate reductase (CoM/CoB). Now EC 1.3.4.1, fumarate reductase (CoM/CoB) (EC 1.3.98.2 created 2014, deleted 2014)]
Accepted name: coproporphyrinogen dehydrogenase
Reaction: coproporphyrinogen III + 2 S-adenosyl-L-methionine = protoporphyrinogen IX + 2 CO2 + 2 L-methionine + 2 5'-deoxyadenosine
For diagram of reaction click here.
Other name(s): oxygen-independent coproporphyrinogen-III oxidase; HemN; coproporphyrinogen III oxidase
Systematic name: coproporphyrinogen-III:S-adenosyl-L-methionine oxidoreductase (decarboxylating)
Comments: This enzyme differs from EC 1.3.3.3, coproporphyrinogen oxidase, by using S-adenosyl-L-methionine (AdoMet) instead of oxygen as oxidant. It occurs mainly in bacteria, whereas eukaryotes use the oxygen-dependent oxidase. The reaction starts by using an electron from the reduced form of the enzyme's [4Fe-4S] cluster to split AdoMet into methionine and the radical 5'-deoxyadenosin-5'-yl. This radical initiates attack on the 2-carboxyethyl groups, leading to their conversion into vinyl groups. This conversion, —·CH-CH2-COO- → —CH=CH2 + CO2 + e- replaces the electron initially used.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Layer, G., Verfürth, K., Mahlitz, E. and Jahn, D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J. Biol. Chem. 277 (2002) 34136-34142. [PMID: 12114526]
2. Layer, G., Moser, J., Heinz, D.W., Jahn, D. and Schubert, W.D. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22 (2003) 6214-6224. [PMID: 14633981]
Accepted name: 5a,11a-dehydrotetracycline reductase
Reaction: tetracycline + oxidized coenzyme F420 = 5a,11a-dehydrotetracycline + reduced coenzyme F420
For diagram of reaction click here.
Other name(s): oxyR (gene name); 12-dehydrotetracycline dehydrogenase; dehydrooxytetracycline dehydrogenase; 12-dehydrotetracycline reductase
Systematic name: tetracycline:coenzyme F420 dehydrogenase
Comments: The enzyme, characterized from the bacteria Streptomyces aureofaciens and Streptomyces rimosus, catalyses the last step in the biosynthesis of the tetracycline antibiotics tetracycline and oxytetracycline.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. McCormick, J.R.D., Hirsch, U., Sjolander, N.O. and Doerschuk, A.P. Cosynthesis of tetracyclines by pairs of Streptomyces aureofaciens mutants. J. Am. Chem. Soc. 82 (1960) 5006-5007.
2. Miller, P.A., Sjolander, N.O., Nalesnyk, S., Arnold, N., Johnson, S., Doerschuk, A.P. and McCormick, J.R.D. Cosynthetic factor I, a factor involved in hydrogen-transfer in Streptomyces aureofaciens. J. Am. Chem. Soc. 82 (1960) 5002-5003.
3. McCormick, J.R.D. and Morton, G.O. Identity of cosynthetic factor I of Streptomyces aureofaciens and fragment FO from coenzyme F420 of Methanobacterium species. J. Am. Chem. Soc. 104 (1982) 4014-4015.
4. Wang, P., Bashiri, G., Gao, X., Sawaya, M.R. and Tang, Y. Uncovering the enzymes that catalyze the final steps in oxytetracycline biosynthesis. J. Am. Chem. Soc. 135 (2013) 7138-7141. [PMID: 23621493]
Accepted name: hydrogen peroxide-dependent heme synthase
Reaction: Fe-coproporphyrin III + 2 H2O2 = protoheme + 2 CO2 + 4 H2O (overall reaction)
(1a) Fe-coproporphyrin III + H2O2 = harderoheme III + CO2 + 2 H2O
(1b) harderoheme III + H2O2 = protoheme + CO2 + 2 H2O
Other name(s): coproheme III oxidative decarboxylase; hemQ (gene name)
Systematic name: Fe-coproporphyrin III:hydrogen peroxide oxidoreductase (decarboxylating)
Comments: The enzyme participates in a heme biosynthesis pathway found in Gram-positive bacteria. The initial decarboxylation step is fast and yields the three-propanoate harderoheme isomer III. The second decarboxylation is much slower. cf. EC 1.3.98.6, SAM-dependent heme synthase.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, PDB, CAS registry number:
References:
1. Dailey, T.A., Boynton, T.O., Albetel, A.N., Gerdes, S., Johnson, M.K. and Dailey, H.A. Discovery and characterization of HemQ: an essential heme biosynthetic pathway component. J. Biol. Chem 285 (2010) 25978-25986. [PMID: 20543190]
2. Celis, A.I., Streit, B.R., Moraski, G.C., Kant, R., Lash, T.D., Lukat-Rodgers, G.S., Rodgers, K.R. and DuBois, J.L. Unusual peroxide-dependent, heme-transforming reaction catalyzed by HemQ. Biochemistry 54 (2015) 4022-4032. [PMID: 26083961]
3. Hofbauer, S., Mlynek, G., Milazzo, L., Puhringer, D., Maresch, D., Schaffner, I., Furtmuller, P.G., Smulevich, G., Djinovic-Carugo, K. and Obinger, C. Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies. FEBS J. 283 (2016) 4386-4401. [PMID: 27758026]
4. Celis, A.I., Gauss, G.H., Streit, B.R., Shisler, K., Moraski, G.C., Rodgers, K.R., Lukat-Rodgers, G.S., Peters, J.W. and DuBois, J.L. Structure-based mechanism for oxidative decarboxylation reactions mediated by amino acids and heme propionates in coproheme decarboxylase (HemQ). J. Am. Chem. Soc. 139 (2017) 1900-1911. [PMID: 27936663]
Accepted name: AdoMet-dependent heme synthase
Reaction: Fe-coproporphyrin III + 2 S-adenosyl-L-methionine = protoheme + 2 CO2 + 2 5'-deoxyadenosine + 2 L-methionine
Other name(s): ahbD (gene name); SAM-dependent heme synthase
Systematic name: Fe-coproporphyrin III:S-adenosyl-L-methionine oxidoreductase (decarboxylating)
Comments: This radical AdoMet enzyme participates in a heme biosynthesis pathway found in archaea and sulfur-reducing bacteria. cf. EC 1.3.98.5, hydrogen peroxide-dependent heme synthase.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Bali, S., Lawrence, A.D., Lobo, S.A., Saraiva, L.M., Golding, B.T., Palmer, D.J., Howard, M.J., Ferguson, S.J. and Warren, M.J. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc. Natl Acad. Sci. USA 108 (2011) 18260-18265. [PMID: 21969545]
2. Kuhner, M., Haufschildt, K., Neumann, A., Storbeck, S., Streif, J. and Layer, G. The alternative route to heme in the methanogenic archaeon Methanosarcina barkeri. Archaea 2014 (2014) 327637. [PMID: 24669201]
Accepted name: [mycofactocin precursor peptide]-tyrosine decarboxylase
Reaction: C-terminal [mycofactocin precursor peptide]-glycyl-L-valyl-L-tyrosine + S-adenosyl-L-methionine = C-terminal [mycofactocin precursor peptide]-glycyl-L-valyl-4-[2-aminoethenyl]phenol + CO2 + 5'-deoxyadenosine + L-methionine
Other name(s): mftC (gene name)
Systematic name: C-terminal [mycofactocin precursor peptide]-glycyl-L-valyl-L-tyrosine L-tyrosine-carboxylyase
Comments: This is a bifunctional radical AdoMet (radical SAM) enzyme that catalyses the first two steps in the biosynthesis of the enzyme cofactor mycofactocin. Activity requires the presence of the MftB chaperone. The other activity of the enzyme is EC 4.1.99.26, 3-amino-5-[(4-hydroxyphenyl)methyl]-4,4-dimethylpyrrolidin-2-one synthase.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Haft, D.H. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics 12 (2011) 21. [PMID: 21223593]
2. Bruender, N.A. and Bandarian, V. The radical S-adenosyl-L-methionine enzyme MftC catalyzes an oxidative decarboxylation of the C-terminus of the MftA peptide. Biochemistry 55 (2016) 2813-2816. [PMID: 27158836]
3. Khaliullin, B., Ayikpoe, R., Tuttle, M. and Latham, J.A. Mechanistic elucidation of the mycofactocin-biosynthetic radical S-adenosylmethionine protein, MftC. J. Biol. Chem. 292 (2017) 13022-13033. [PMID: 28634235]
4. Ayikpoe, R., Ngendahimana, T., Langton, M., Bonitatibus, S., Walker, L.M., Eaton, S.S., Eaton, G.R., Pandelia, M.E., Elliott, S.J. and Latham, J.A. Spectroscopic and electrochemical characterization of the mycofactocin biosynthetic protein, MftC, provides insight into its redox flipping mechanism. Biochemistry 58 (2019) 940-950. [PMID: 30628436]
[EC 1.3.99.2 Transferred entry: butyryl-CoA dehydrogenase. Now EC 1.3.8.1, butyryl-CoA dehydrogenase. (EC 1.3.99.2 created 1961 as EC 1.3.2.1, transferred 1964 to EC 1.3.99.2, deleted 2011)]
[EC 1.3.99.3 Transferred entry: acyl-CoA dehydrogenase, now EC 1.3.8.7, medium-chain acyl-CoA dehydrogenase, EC 1.3.8.8, long-chain acyl-CoA dehydrogenase and EC 1.3.8.9, very-long-chain acyl-CoA dehydrogenase (EC 1.3.99.3 created 1961 as EC 1.3.2.2, transferred 1964 to EC 1.3.99.3, deleted 2012)]
Accepted name: 3-oxosteroid 1-dehydrogenase
Reaction: a 3-oxosteroid + acceptor = a 3-oxo-δ1-steroid + reduced acceptor
Other name(s): 3-oxosteroid δ1-dehydrogenase; δ1-dehydrogenase; 3-ketosteroid-1-en-dehydrogenase; 3-ketosteroid-δ1-dehydrogenase; 1-ene-dehydrogenase; 3-oxosteroid:(2,6-dichlorphenolindophenol) δ1-oxidoreductase; 4-en-3-oxosteroid:(acceptor)-1-en-oxido-reductase; δ1-steroid reductase; 3-oxosteroid:(acceptor) δ1-oxidoreductase
Systematic name: 3-oxosteroid:acceptor δ1-oxidoreductase
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9029-04-3
References:
1. Levy, H.R. and Talalay, P. Bacterial oxidation of steroids. II. Studies on the enzymatic mechanisms of ring A dehydrogenation. J. Biol. Chem. 234 (1959) 2014-2021.
Accepted name: 3-oxo-5α-steroid 4-dehydrogenase (acceptor)
Reaction: a 3-oxo-5α-steroid + acceptor = a 3-oxo-Δ4-steroid + reduced acceptor
Other name(s): steroid 5α-reductase; 3-oxosteroid Δ4-dehydrogenase; 3-oxo-5α-steroid Δ4-dehydrogenase; steroid Δ4-5α-reductase; Δ4-3-keto steroid 5α-reductase; Δ4-3-oxo steroid reductase; Δ4-3-ketosteroid5α-oxidoreductase; Δ4-3-oxosteroid-5α-reductase; 3-keto-Δ4-steroid-5α-reductase; 5α-reductase; testosterone 5α-reductase; 4-ene-3-ketosteroid-5α-oxidoreductase; Δ4-5α-dehydrogenase; 3-oxo-5α-steroid:(acceptor) Δ4-oxidoreductase; tesI (gene name)
Systematic name: 3-oxo-5α-steroid:acceptor Δ4-oxidoreductase
Comments: A flavoprotein. This bacterial enzyme, characterized from Comamonas testosteroni, is involved in androsterone degradation. cf. EC 1.3.1.22, 3-oxo-5α-steroid 4-dehydrogenase (NADP+).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 9036-43-5
References:
1. Levy, H.R. and Talalay, P. Bacterial oxidation of steroids. II. Studies on the enzymatic mechanisms of ring A dehydrogenation. J. Biol. Chem. 234 (1959) 2014-2021. [PMID: 13673006]
2. Florin, C., Kohler, T., Grandguillot, M. and Plesiat, P. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: gene and protein characterization. J. Bacteriol. 178 (1996) 3322-3330. [PMID: 8655514]
3. Horinouchi, M., Hayashi, T., Yamamoto, T. and Kudo, T. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 69 (2003) 4421-4430. [PMID: 12902225]
Accepted name: 3-oxo-5β-steroid 4-dehydrogenase
Reaction: a 3-oxo-5β-steroid + acceptor = a 3-oxo-Δ4-steroid + reduced acceptor
Other name(s): 3-oxo-5β-steroid:(acceptor) Δ4-oxidoreductase
Systematic name: 3-oxo-5β-steroid:acceptor Δ4-oxidoreductase
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 9067-97-4
References:
1. Davidson, S.J. and Talalay, P. Purification and mechanism of action of a steroid δ4-5β-dehydrogenase. J. Biol. Chem. 241 (1966) 906-915. [PMID: 5907467]
[EC 1.3.99.7 Transferred entry: glutaryl-CoA dehydrogenase. Now EC 1.3.8.6, glutaryl-CoA dehydrogenase (EC 1.3.99.7 created 1972, deleted 2012)]
Accepted name: 2-furoyl-CoA dehydrogenase
Reaction: 2-furoyl-CoA + H2O + acceptor = S-(5-hydroxy-2-furoyl)-CoA + reduced acceptor
Other name(s): furoyl-CoA hydroxylase; 2-furoyl coenzyme A hydroxylase; 2-furoyl coenzyme A dehydrogenase; 2-furoyl-CoA:(acceptor) 5-oxidoreductase (hydroxylating)
Systematic name: 2-furoyl-CoA:acceptor 5-oxidoreductase (hydroxylating)
Comments: A copper protein. The oxygen atom of the -OH produced is derived from water, not O2; the actual oxidative step is probably dehydrogenation of a hydrated form -CHOH-CH2- to -C(OH)=CH-, which tautomerizes non-enzymically to -CO-CH2-, giving (5-oxo-4,5-dihydro-2-furoyl)-CoA. Methylene blue, nitro blue, tetrazolium and a membrane fraction from Pseudomonas putida can act as acceptors.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, Metacyc, CAS registry number: 9068-18-2
References:
1. Kitcher, J.P., Trudgill, P.W. and Rees, J.S. Purification and properties of 2-furoyl-coenzyme A hydroxylase from Pseudomonas putida F2. Biochem. J. 130 (1972) 121-132.
[EC 1.3.99.9 Transferred entry: now EC 1.21.99.1, β-cyclopiazonate dehydrogenase (EC 1.3.99.9 created 1976, deleted 2002)]
[EC 1.3.99.10 Transferred entry: isovaleryl-CoA dehydrogenase. Now EC 1.3.8.4, isovaleryl-CoA dehydrogenase (EC 1.3.99.10 created 1978, modified 1986, deleted 2012)]
[EC 1.3.99.11 Transferred entry: dihydroorotate dehydrogenase. As the acceptor is now known, the enzyme has been transferred to EC 1.3.5.2, dihydroorotate dehydrogenase (EC 1.3.99.11 created 1983, deleted 2009)]
[EC 1.3.99.12 Transferred entry: 2-methylacyl-CoA dehydrogenase. Now classified as EC 1.3.8.5, 2-methyl-branched-chain-enoyl-CoA reductase. (EC 1.3.99.12 created 1986, deleted 2020)]
[EC 1.3.99.13 Transferred entry: long-chain-acyl-CoA dehydrogenase. Now EC 1.3.8.8, long-chain-acyl-CoA dehydrogenase (EC 1.3.99.13 created 1989, deleted 2012)]
Accepted name: cyclohexanone dehydrogenase
Reaction: cyclohexanone + acceptor = cyclohex-2-enone + reduced acceptor
Other name(s): cyclohexanone:(acceptor) 2-oxidoreductase
Systematic name: cyclohexanone:acceptor 2-oxidoreductase
Comments: 2,6-Dichloroindophenol can act as acceptor. The corresponding ketones of cyclopentane and cycloheptane cannot act as donors.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 123516-43-8
References:
1. Dangel, W., Tschech, A. and Fuchs, G. Enzyme reactions involved in anaerobic cyclohexanol metabolism by a denitrifying Pseudomonas species. Arch. Microbiol. 152 (1989) 273-279. [PMID: 2505723]
[EC 1.3.99.15 Transferred entry: benzoyl-CoA reductase. Now EC 1.3.7.8. (EC 1.3.99.15 created 1999, deleted 2011)]
Accepted name: isoquinoline 1-oxidoreductase
Reaction: isoquinoline + acceptor + H2O = isoquinolin-1(2H)-one + reduced acceptor
Systematic name: isoquinoline:acceptor 1-oxidoreductase (hydroxylating)
Comments: the enzyme from Pseudomonas diminuta is specific towards N-containing N-heterocyclic substrates, including isoquinoline, isoquinolin-5-ol, phthalazine and quinazoline. Electron acceptors include 1,2-benzoquinone, cytochrome c, ferricyanide, iodonitrotetrazolium chloride, nitroblue tetrazolium, Meldola blue and phenazine methosulfate.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 155948-73-5
References:
1. Lehmann, M., Tshisuaka, B., Fetzner, S., Roger, P. and Lingens, F. Purification and characterization of isoquinoline 1-oxidoreductase from Pseudomonas diminuta 7, a novel molybdenum-containing hydroxylase. J. Biol. Chem. 269 (1994) 11254-11260. [PMID: 8157655]
2. Lehmann, M., Tshisuaka, B., Fetzner, S. and Lingens, F. Molecular cloning of the isoquinoline 1-oxidoreductase genes from Pseudomonas diminuta 7, structural analysis of iorA and iorB, and sequence comparisons with other molybdenum-containing hydroxylases. J. Biol. Chem. 270 (1995) 14420-14429. [PMID: 7782304]
Accepted name: quinoline 2-oxidoreductase
Reaction: quinoline + acceptor + H2O = quinolin-2(1H)-one + reduced acceptor
Systematic name: quinoline:acceptor 2-oxidoreductase (hydroxylating)
Comments: Quinolin-2-ol, quinolin-7-ol, quinolin-8-ol, 3-, 4- and 8-methylquinolines and 8-chloroquinoline are substrates. Iodonitrotetrazolium chloride can act as an electron acceptor.
Links to other databases: BRENDA, EAWAG-BBD, EXPASY, KEGG, Metacyc, PDB, CAS registry number: 132264-32-5
References:
1. Bauder, R., Tshisuaka, B. and Lingens, F. Microbial metabolism of quinoline and related compounds. VII. Quinoline oxidoreductase from Pseudomonas putida: a molybdenum-containing enzyme. Biol. Chem. Hoppe-Seyler 371 (1990) 1137-1144.
2. Tshisuaka, B., Kappl, R., Huttermann, J. and Lingens, F. Quinoline oxidoreductase from Pseudomonas putida 86: an improved purification procedure and electron paramagnetic resonance spectroscopy. Biochemistry 32 (1993) 12928-12934. [PMID: 8251516]
3. Peschke, B. and Lingens, F. Microbial metabolism of quinoline and related compounds. XII. Isolation and characterization of the quinoline oxidoreductase from Rhodococcus sp. B1 compared with the quinoline oxidoreductase from Pseudomonas putida 86. Biol. Chem. Hoppe-Seyler 372 (1991) 1081-1088. [PMID: 1789933]
4. Schach, S., Tshisuaka, B., Fetzner, S. and Lingens, F. Quinoline 2-oxidoreductase and 2-oxo-1,2-dihydroquinoline 5,6-dioxygenase from Comamonas testosteroni 63. The first two enzymes in quinoline and 3-methylquinoline degradation. Eur. J. Biochem. 232 (1995) 536-544. [PMID: 7556204]
Accepted name: quinaldate 4-oxidoreductase
Reaction: quinaldate + acceptor + H2O = kynurenate + reduced acceptor
Other name(s): quinaldic acid 4-oxidoreductase
Systematic name: quinoline-2-carboxylate:acceptor 4-oxidoreductase (hydroxylating)
Comments: the enzyme from Pseudomonas sp. AK2 also acts on quinoline-8-carboxylate, whereas that from Serratia marcescens 2CC-1 will oxidize nicotinate; quinaldate is a substrate for both of these enzymes. 2,4,6-Trinitrobenzene sulfonate, 1,4-benzoquinone, 1,2-naphthoquinone, nitroblue tetrazolium, thionine and menadione will serve as an electron acceptor for the former enzyme and ferricyanide for the latter; Meldola blue, iodonitrotetrazolium chloride, phenazine methosulfate, 2,6-dichlorophenolindophenol and cytochrome c will act as electron acceptors for both.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 149885-77-8
References:
1. Sauter, M., Tshisuaka, B., Fetzner, S. and Lingens, F. Microbial metabolism of quinoline and related compounds. XX. Quinaldic acid 4-oxidoreductase from Pseudomonas sp. AK-2 compared to other procaryotic molybdenum-containing hydroxylases. Biol. Chem. Hoppe Seyler 374 (1993) 1037-1046. [PMID: 8292263]
2. Fetzner, S. and Lingens, F. Microbial metabolism of quinoline and related compounds. XVIII. Purification and some properties of the molybdenum- and iron-containing quinaldic acid 4-oxidoreductase from Serratia marcescens 2CC-1. Biol. Chem. Hoppe-Seyler 374 (1993) 363-376. [PMID: 8357532]
Accepted name: quinoline-4-carboxylate 2-oxidoreductase
Reaction: quinoline-4-carboxylate + acceptor + H2O = 2-oxo-1,2-dihydroquinoline-4-carboxylate + reduced acceptor
For diagram, click here
Other name(s): quinaldic acid 4-oxidoreductase; quinoline-4-carboxylate:acceptor 2-oxidoreductase (hydroxylating)
Systematic name: quinoline-4-carboxylate:acceptor 2-oxidoreductase (hydroxylating)
Comments: A molybdenum—iron—sulfur flavoprotein with molybdopterin cytosine dinucleotide as the molybdenum cofactor. Quinoline, 4-methylquinoline and 4-chloroquinoline can also serve as substrates for the enzyme from Agrobacterium sp. 1B. Iodonitrotetrazolium chloride, thionine, menadione and 2,6-dichlorophenolindophenol can act as electron acceptors.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 175780-18-4
References:
1. Bauer, G. and Lingens, F. Microbial metabolism of quinoline and related compounds. XV. Quinoline-4-carboxylic acid oxidoreductase from Agrobacterium spec.1B: a molybdenum-containing enzyme. Biol. Chem. Hoppe-Seyler 373 (1992) 699-705. [PMID: 1418685]
[EC 1.3.99.20 Transferred entry: EC 1.3.99.20, 4-hydroxybenzoyl-CoA reductase. Now EC 1.3.7.9, 4-hydroxybenzoyl-CoA reductase. (EC 1.3.99.20 created 2000, deleted 2011)]
[EC 1.3.99.21 Transferred entry: (R)-benzylsuccinyl-CoA dehydrogenase. Now EC 1.3.8.3, (R)-benzylsuccinyl-CoA dehydrogenase (EC 1.3.99.21 created 2003 as EC 1.3.99.21, deleted 2012)]
[EC 1.3.99.22 Transferred entry: coproporphyrinogen dehydrogenase. Now EC 1.3.98.3, coproporphyrinogen dehydrogenase (EC 1.3.99.22 created 2004, deleted 2016)]
Accepted name: all-trans-retinol 13,14-reductase
Reaction: all-trans-13,14-dihydroretinol + acceptor = all-trans-retinol + reduced acceptor
For diagram of reaction click here.
Other name(s): retinol saturase; RetSat; (13,14)-all-trans-retinol saturase; all-trans-retinol:all-trans-13,14-dihydroretinol saturase
Systematic name: all-trans-13,14-dihydroretinol:acceptor 13,14-oxidoreductase
Comments: The reaction is only known to occur in the opposite direction to that given above, with the enzyme being specific for all-trans-retinol as substrate. Neither all-trans-retinoic acid nor 9-cis, 11-cis or 13-cis-retinol isomers are substrates. May play a role in the metabolism of vitamin A.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number: 879291-21-1
References:
1. Moise, A.R., Kuksa, V., Imanishi, Y. and Palczewski, K. Identification of all-trans-retinol:all-trans-13,14-dihydroretinol saturase. J. Biol. Chem. 279 (2004) 50230-50242. [PMID: 15358783]
[EC 1.3.99.24 Transferred entry: 2-amino-4-deoxychorismate dehydrogenase. Now EC 1.3.8.16, 2-amino-4-deoxychorismate dehydrogenase (EC 1.3.99.24 created 2008, deleted 2020)]
Accepted name: carvone reductase
Reaction: (1) (+)-dihydrocarvone + acceptor = (–)-carvone + reduced acceptor
(2) (–)-isodihydrocarvone + acceptor = (+)-carvone + reduced acceptor
For diagram of reaction click here.
Glossary: (+)-dihydrocarvone = (1S,4R)-menth-8-en-2-one
(+)-isodihydrocarvone = (1S,4R)-menth-8-en-2-one
(–)-carvone = (4R)-mentha-1(6),8-dien-6-one = (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one
Systematic name: (+)-dihydrocarvone:acceptor 1,6-oxidoreductase
Comments: This enzyme participates in the carveol and dihydrocarveol degradation pathway of the Gram-positive bacterium Rhodococcus erythropolis DCL14. The enzyme has not been purified, and requires an unknown cofactor, which is different from NAD+, NADP+ or a flavin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. van der Werf, M.J. and Boot, A.M. Metabolism of carveol and dihydrocarveol in Rhodococcus erythropolis DCL14. Microbiology 146 (2000) 1129-1141. [PMID: 10832640]
Accepted name: all-trans-ζ-carotene desaturase
Reaction: all-trans-ζ-carotene + 2 acceptor = all-trans-lycopene + 2 reduced acceptor (overall reaction)
(1a) all-trans-ζ-carotene + acceptor = all-trans-neurosporene + reduced acceptor
(1b) all-trans-neurosporene + acceptor = all-trans-lycopene + reduced acceptor
For diagram of reaction click here.
Other name(s): CrtIb; phytoene desaturase (ambiguous); 2-step phytoene desaturase (ambiguous); two-step phytoene desaturase (ambiguous); CrtI (ambiguous)
Systematic name: all-trans-ζ-carotene:acceptor oxidoreductase
Comments: This enzyme is involved in carotenoid biosynthesis.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Iniesta, A.A., Cervantes, M. and Murillo, F.J. Cooperation of two carotene desaturases in the production of lycopene in Myxococcus xanthus. FEBS J. 274 (2007) 4306-4314. [PMID: 17662111]
Accepted name: 1-hydroxycarotenoid 3,4-desaturase
Reaction: 1-hydroxy-1,2-dihydrolycopene + acceptor = 1-hydroxy-3,4-didehydro-1,2-dihydrolycopene + reduced acceptor
For diagram of reaction click here or click here
Other name(s): CrtD; hydroxyneurosporene desaturase; carotenoid 3,4-dehydrogenase; 1-hydroxy-carotenoid 3,4-dehydrogenase
Systematic name: 1-hydroxy-1,2-dihydrolycopene:acceptor oxidoreductase
Comments: The enzymes from Rubrivivax gelatinosus and Rhodobacter sphaeroides prefer the acyclic carotenoids (e.g. 1-hydroxy-1,2-dihydroneurosporene, 1-hydroxy-1,2-dihydrolycopene) as substrates. The conversion rate for the 3,4-desaturation of the monocyclic 1'-hydroxy-1',2'-dihydro-γ-carotene is lower [2,3]. The enzyme from the marine bacterium strain P99-3 shows high activity with the monocyclic carotenoid 1'-hydroxy-1',2'-dihydro-γ-carotene [1]. The enzyme from Rhodobacter sphaeroides utilizes molecular oxygen as the electron acceptor in vitro [3]. However, oxygen is unlikely to be the natural electron acceptor under anaerobic conditions.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Teramoto, M., Rahlert, N., Misawa, N. and Sandmann, G. 1-Hydroxy monocyclic carotenoid 3,4-dehydrogenase from a marine bacterium that produces myxol. FEBS Lett. 570 (2004) 184-188. [PMID: 15251462]
2. Steiger, S., Astier, C. and Sandmann, G. Substrate specificity of the expressed carotenoid 3,4-desaturase from Rubrivivax gelatinosus reveals the detailed reaction sequence to spheroidene and spirilloxanthin. Biochem. J. 349 (2000) 635-640. [PMID: 10880364]
3. Albrecht, M., Ruther, A. and Sandmann, G. Purification and biochemical characterization of a hydroxyneurosporene desaturase involved in the biosynthetic pathway of the carotenoid spheroidene in Rhodobacter sphaeroides. J. Bacteriol. 179 (1997) 7462-7467. [PMID: 9393712]
Accepted name: phytoene desaturase (neurosporene-forming)
Reaction: 15-cis-phytoene + 3 acceptor = all-trans-neurosporene + 3 reduced acceptor (overall reaction)
(1a) 15-cis-phytoene + acceptor = all-trans-phytofluene + reduced acceptor
(1b) all-trans-phytofluene + acceptor = all-trans-ζ-carotene + reduced acceptor
(1c) all-trans-ζ-carotene + acceptor = all-trans-neurosporene + reduced acceptor
For diagram of reaction click here.
Other name(s): 3-step phytoene desaturase; three-step phytoene desaturase; phytoene desaturase (ambiguous); CrtI (ambiguous)
Systematic name: 15-cis-phytoene:acceptor oxidoreductase (neurosporene-forming)
Comments: This enzyme is involved in carotenoid biosynthesis and catalyses up to three desaturation steps (cf. EC 1.3.99.29 [phytoene desaturase (ζ-carotene-forming)], EC 1.3.99.30 [phytoene desaturase (3,4-didehydrolycopene-forming)], EC 1.3.99.31 [phytoene desaturase (lycopene-forming)]). The enzyme is activated by FAD. NAD+, NADP+ or ATP show no activating effect [1].
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Raisig, A., Bartley, G., Scolnik, P. and Sandmann, G. Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus overexpressed in Escherichia coli. J. Biochem. 119 (1996) 559-564. [PMID: 8830054]
2. Wang, C.W. and Liao, J.C. Alteration of product specificity of Rhodobacter sphaeroides phytoene desaturase by directed evolution. J. Biol. Chem. 276 (2001) 41161-41164. [PMID: 11526111]
Accepted name: phytoene desaturase (ζ-carotene-forming)
Reaction: 15-cis-phytoene + 2 acceptor = all-trans-ζ-carotene + 2 reduced acceptor (overall reaction)
(1a) 15-cis-phytoene + acceptor = all-trans-phytofluene + reduced acceptor
(1b) all-trans-phytofluene + acceptor = all-trans-ζ-carotene + reduced acceptor
For diagram of reaction click here.
Other name(s): CrtIa; 2-step phytoene desaturase (ambiguous); two-step phytoene desaturase (ambiguous)
Systematic name: 15-cis-phytoene:acceptor oxidoreductase (ζ-carotene-forming)
Comments: The enzyme is involved in carotenoid biosynthesis and catalyses up to two desaturation steps (cf. EC 1.3.99.28 [phytoene desaturase (neurosporene-forming)], EC 1.3.99.30 [phytoene desaturase (3,4-didehydrolycopene-forming)] and EC 1.3.99.31 [phytoene desaturase (lycopene-forming)]).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Iniesta, A.A., Cervantes, M. and Murillo, F.J. Cooperation of two carotene desaturases in the production of lycopene in Myxococcus xanthus. FEBS J. 274 (2007) 4306-4314. [PMID: 17662111]
Accepted name: phytoene desaturase (3,4-didehydrolycopene-forming)
Reaction: 15-cis-phytoene + 5 acceptor = all-trans-3,4-didehydrolycopene + 5 reduced acceptor (overall reaction)
(1a) 15-cis-phytoene + acceptor = all-trans-phytofluene + reduced acceptor
(1b) all-trans-phytofluene + acceptor = all-trans-ζ-carotene + reduced acceptor
(1c) all-trans-ζ-carotene + acceptor = all-trans-neurosporene + reduced acceptor
(1d) all-trans-neurosporene + acceptor = all-trans-lycopene + reduced acceptor
(1e) all-trans-lycopene + acceptor = all-trans-3,4-didehydrolycopene + reduced acceptor
For diagram of reaction click here.
Other name(s): 5-step phytoene desaturase; five-step phytoene desaturase; phytoene desaturase (ambiguous); Al-1
Systematic name: 15-cis-phytoene:acceptor oxidoreductase (3,4-didehydrolycopene-forming)
Comments: This enzyme is involved in carotenoid biosynthesis and catalyses up to five desaturation steps (cf. EC 1.3.99.28 [phytoene desaturase (neurosporene-forming)], EC 1.3.99.29 [phytoene desaturase (ζ-carotene-forming)] and EC 1.3.99.31 [phytoene desaturase (lycopene-forming)]).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Hausmann, A. and Sandmann, G. A single five-step desaturase is involved in the carotenoid biosynthesis pathway to β-carotene and torulene in Neurospora crassa. Fungal Genet. Biol. 30 (2000) 147-153. [PMID: 11017770]
2. Estrada, A.F., Maier, D., Scherzinger, D., Avalos, J. and Al-Babili, S. Novel apocarotenoid intermediates in Neurospora crassa mutants imply a new biosynthetic reaction sequence leading to neurosporaxanthin formation. Fungal Genet. Biol. 45 (2008) 1497-1505. [PMID: 18812228]
Accepted name: phytoene desaturase (lycopene-forming)
Reaction: 15-cis-phytoene + 4 acceptor = all-trans-lycopene + 4 reduced acceptor (overall reaction)
(1a) 15-cis-phytoene + acceptor = all-trans-phytofluene + reduced acceptor
(1b) all-trans-phytofluene + acceptor = all-trans-ζ-carotene + reduced acceptor
(1c) all-trans-ζ-carotene + acceptor = all-trans-neurosporene + reduced acceptor
(1d) all-trans-neurosporene + acceptor = all-trans-lycopene + reduced acceptor
For diagram of reaction click here.
Other name(s): 4-step phytoene desaturase; four-step phytoene desaturase; phytoene desaturase (ambiguous); CrtI (ambiguous)
Systematic name: 15-cis-phytoene:acceptor oxidoreductase (lycopene-forming)
Comments: Requires FAD. The enzyme is involved in carotenoid biosynthesis and catalyses up to four desaturation steps (cf. EC 1.3.99.28 [phytoene desaturase (neurosporene-forming)], EC 1.3.99.29 [phytoene desaturase (ζ-carotene-forming)] and EC 1.3.99.30 [phytoene desaturase (3,4-didehydrolycopene-forming)]).
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Fraser, P.D., Misawa, N., Linden, H., Yamano, S., Kobayashi, K. and Sandmann, G. Expression in Escherichia coli, purification, and reactivation of the recombinant Erwinia uredovora phytoene desaturase. J. Biol. Chem. 267 (1992) 19891-19895. [PMID: 1400305]
Accepted name: glutaryl-CoA dehydrogenase (non-decarboxylating)
Reaction: glutaryl-CoA + acceptor = (E)-glutaconyl-CoA + reduced acceptor
Glossary: (E)-glutaconyl-CoA = (2E)-4-carboxybut-2-enoyl-CoA
Other name(s): GDHDes; nondecarboxylating glutaryl-coenzyme A dehydrogenase; nondecarboxylating glutaconyl-coenzyme A-forming GDH
Systematic name: glutaryl-CoA:acceptor 2,3-oxidoreductase (non-decarboxylating)
Comments: The enzyme contains FAD. The anaerobic, sulfate-reducing bacterium Desulfococcus multivorans contains two glutaryl-CoA dehydrogenases: a decarboxylating enzyme (EC 1.3.8.6), and a nondecarboxylating enzyme (this entry). The two enzymes cause different structural changes around the glutaconyl carboxylate group, primarily due to the presence of either a tyrosine or a valine residue, respectively, at the active site.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Wischgoll, S., Taubert, M., Peters, F., Jehmlich, N., von Bergen, M. and Boll, M. Decarboxylating and nondecarboxylating glutaryl-coenzyme A dehydrogenases in the aromatic metabolism of obligately anaerobic bacteria. J. Bacteriol. 191 (2009) 4401-4409. [PMID: 19395484]
2. Wischgoll, S., Demmer, U., Warkentin, E., Gunther, R., Boll, M. and Ermler, U. Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme A dehydrogenases. Biochemistry 49 (2010) 5350-5357. [PMID: 20486657]
Accepted name: urocanate reductase
Reaction: dihydrourocanate + acceptor = urocanate + reduced acceptor
For diagram of reaction click here.
Glossary: urocanate = 3-(1H-imidazol-4-yl)prop-2-enoate
dihydrourocanate = 3-(1H-imidazol-4-yl)propanoate
Other name(s): urdA (gene name)
Systematic name: dihydrourocanate:acceptor oxidoreductase
Comments: The enzyme from the bacterium Shewanella oneidensis MR-1 contains a noncovalently-bound FAD and a covalently-bound FMN. It functions as part of an anaerobic electron transfer chain that utilizes urocanate as the terminal electron acceptor. The activity has been demonstrated with the artificial donor reduced methyl viologen.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Bogachev, A.V., Bertsova, Y.V., Bloch, D.A. and Verkhovsky, M.I. Urocanate reductase: identification of a novel anaerobic respiratory pathway in Shewanella oneidensis MR-1. Mol. Microbiol. 86 (2012) 1452-1463. [PMID: 23078170]
[EC 1.3.99.34 Transferred entry: 2,3-bis-O-geranylgeranyl-sn-glycerol 1-phosphate reductase (donor). Now classified as EC 1.3.7.11, 2,3-bis-O-geranylgeranyl-sn-glycero-phospholipid reductase. (EC 1.3.99.34 created 2013, deleted 2015)]
[EC 1.3.99.35 Transferred entry: chlorophyllide a reductase. Now EC 1.3.7.15, chlorophyllide a reductase (EC 1.3.99.35 created 2014, deleted 2016)]
Accepted name: cypemycin cysteine dehydrogenase (decarboxylating)
Reaction: cypemycin(1-18)-L-Cys-L-Leu-L-Val-L-Cys + acceptor = C3.19,S21-cyclocypemycin(1-18)-L-Ala-L-Leu-N-thioethenyl-L-valinamide + CO2 + H2S + reduced acceptor
For diagram of reaction click here.
Glossary: C3.19,S21-cyclocypemycin(1-18)-L-Ala-L-Leu-N-thioethenyl-L-valinamide = 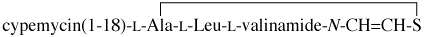
Other name(s): cypemycin decarboxylase; CypD
Systematic name: cypemycin(1-18)-L-Cys-L-Leu-L-Val-L-Cys:acceptor oxidoreductase (decarboxylating)
Comments: Cypemycin, isolated from the bacterium Streptomyces sp. OH-4156, is a peptide antibiotic, member of the linaridins, a class of posttranslationally modified ribosomally synthesized peptides. The enzyme decarboxylates and reduces the C-terminal L-cysteine residue, producing a reactive ethenethiol group that reacts with a dethiolated cysteine upstream to form an aminovinyl-methyl-cysteine loop that is important for the antibiotic activity of the mature peptide.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, PDB, CAS registry number:
References:
1. Claesen, J. and Bibb, M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc. Natl. Acad. Sci. USA 107 (2010) 16297-16302. [PMID: 20805503]
Accepted name: 1-hydroxy-2-isopentenylcarotenoid 3,4-desaturase
Reaction: (1) dihydroisopentenyldehydrorhodopin + acceptor = isopentenyldehydrorhodopin + reduced acceptor
(2) dihydrobisanhydrobacterioruberin + acceptor = bisanhydrobacterioruberin + reduced acceptor
For diagram of reaction click here.
Glossary: bisanhydrobacterioruberin = (2S,2S')-2,2'-bis(3-methylbut-2-en-1-yl)-3,4-didehydro-1,1',2,2'-tetrahydro-ψ,ψ-carotene-1,1'-diol
dihydrobisanhydrobacterioruberin = (2S,2S')-2,2'-bis(3-methylbut-2-en-1-yl)-3,3',4,4'-tetradehydro-1,1',2,2'-tetrahydro-ψ,ψ-carotene-1,1'-diol
dihydroisopentenyldehydrorhodopin = (2S)-2-(3-methylbut-2-en-1-yl)-3,4-didehydro-1,2-dihydro-ψ,ψ-caroten-1-ol
isopentenyldehydrorhodopin = (2S)-2-(3-methylbut-2-en-1-yl)-1,2-dihydro-ψ,ψ-caroten-1-ol
Other name(s): crtD (gene name)
Systematic name: dihydroisopentenyldehydrorhodopin:acceptor 3,4-oxidoreductase
Comments: The enzyme, isolated from the archaeon Haloarcula japonica, is involved in the biosynthesis of the C50 carotenoid bacterioruberin. In this pathway it catalyses the desaturation of the C-3,4 double bond in dihydroisopentenyldehydrorhodopin and the desaturation of the C-3',4' double bond in dihydrobisanhydrobacterioruberin.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Yang, Y., Yatsunami, R., Ando, A., Miyoko, N., Fukui, T., Takaichi, S. and Nakamura, S. Complete biosynthetic pathway of the C50 carotenoid bacterioruberin from lycopene in the extremely halophilic archaeon Haloarcula japonica. J. Bacteriol. 197 (2015) 1614-1623. [PMID: 25712483]
Accepted name: menaquinone-9 β-reductase
Reaction: menaquinone-9 + reduced acceptor = β-dihydromenaquinone-9 + acceptor
For diagram of reaction click here.
Glossary: β-dihydromenaquinone-9 = MK-9(II-H2) = 2-methyl-3-[(2E,10E,14E,18E,22E,26E,30E,33E)-3,7,11,15,19,23,27,31,35-nonamethylhexatriaconta-2,10,14,18,22,26,30,33-octaen-1-yl]naphthalene-1,4-dione
Other name(s): MenJ
Systematic name: menaquinone-9 oxidoreductase (β-dihydromenaquinone-9-forming)
Comments: The enzyme from the bacterium Mycobacterium tuberculosis reduces the β-isoprene unit of menaquinone-9, forming the predominant form of menaquinone found in mycobacteria. Contains FAD.
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Upadhyay, A., Fontes, F.L., Gonzalez-Juarrero, M., McNeil, M.R., Crans, D.C., Jackson, M. and Crick, D.C. Partial saturation of menaquinone in Mycobacterium tuberculosis: function and essentiality of a novel reductase, MenJ. ACS Cent. Sci. 1 (2015) 292-302. [PMID: 26436137]
Accepted name: carotenoid φ-ring synthase
Reaction: carotenoid β-end group + 2 acceptor = carotenoid φ-end group + 2 reduced acceptor
For diagram of reaction click here.
Glossary: chlorobactene = φ,ψ-carotene
β-isorenieratene = φ,β-carotene
isorenieratene = φ,φ-carotene
Other name(s): crtU (gene name) (ambiguous)
Systematic name: carotenoid β-ring:acceptor oxidoreductase/methyltranferase (φ-ring forming)
Comments: The enzyme, found in green sulfur bacteria, some cyanobacteria and some actinobacteria, introduces additional double bonds to the carotenoid β-end group, leading to aromatization of the ionone ring. As a result, one of the methyl groups at C-1 is transferred to position C-2. It is involved in the biosynthesis of carotenoids with φ-type aromatic end groups such as chlorobactene, β-isorenieratene, and isorenieratene.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Moshier, S.E. and Chapman, D.J. Biosynthetic studies on aromatic carotenoids. Biosynthesis of chlorobactene. Biochem. J. 136 (1973) 395-404. [PMID: 4774401]
2. Krugel, H., Krubasik, P., Weber, K., Saluz, H.P. and Sandmann, G. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim. Biophys. Acta 1439 (1999) 57-64. [PMID: 10395965]
3. Frigaard, N.U., Maresca, J.A., Yunker, C.E., Jones, A.D. and Bryant, D.A. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 186 (2004) 5210-5220. [PMID: 15292122]
Accepted name: carotenoid χ-ring synthase
Reaction: carotenoid β-end group + 2 acceptor = carotenoid χ-end group + 2 reduced acceptor
For diagram of reaction click here.
Glossary: okenone = 1'-methoxy-1',2'-dihydro-χ,ψ-caroten-4'-one
renierapurpurin = χ,χ-carotene
synechoxanthin = χ,χ-caroten-18,18'-dioate
Other name(s): crtU (gene name) (ambiguous); cruE (gene name)
Systematic name: carotenoid β-ring:acceptor oxidoreductase/methyltranferase (χ-ring forming)
Comments: The enzyme, found in purple sulfur bacteria (Chromatiaceae) and some cyanobacteria, is involved in the biosynthesis of carotenoids that contain χ-type end groups, such as okenone, renierapurpurin, and synechoxanthin.
Links to other databases: BRENDA, EXPASY, KEGG, Metacyc, CAS registry number:
References:
1. Graham, J.E. and Bryant, D.A. The Biosynthetic pathway for synechoxanthin, an aromatic carotenoid synthesized by the euryhaline, unicellular cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 190 (2008) 7966-7974. [PMID: 18849428]
2. Vogl, K. and Bryant, D.A. Biosynthesis of the biomarker okenone: χ-ring formation. Geobiology 10 (2012) 205-215. [PMID: 22070388]
Accepted name: 3-(methylsulfanyl)propanoyl-CoA 2-dehydrogenase
Reaction: 3-(methylsulfanyl)propanoyl-CoA + acceptor = 3-(methylsulfanyl)acryloyl-CoA + reduced acceptor
Other name(s): dmdC (gene name)
Systematic name: 3-(methylsulfanyl)propanoyl-CoA:acceptor 2-oxidoreductase
Comments: The enzyme, found in marine bacteria, participates in a 3-(methylsulfanyl)propanoate degradation pathway. Based on similar enzymes, the in vivo electron acceptor is likely electron-transfer flavoprotein (ETF).
Links to other databases: BRENDA, EXPASY, KEGG, MetaCyc, CAS registry number:
References:
1. Reisch, C.R., Stoudemayer, M.J., Varaljay, V.A., Amster, I.J., Moran, M.A. and Whitman, W.B. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473 (2011) 208-211. [PMID: 21562561]
2. Bullock, H.A., Luo, H. and Whitman, W.B. Evolution of dimethylsulfoniopropionate metabolism in marine phytoplankton and bacteria. Front. Microbiol. 8 (2017) 637. [PMID: 28469605]
3. Shao, X., Cao, H.Y., Zhao, F., Peng, M., Wang, P., Li, C.Y., Shi, W.L., Wei, T.D., Yuan, Z., Zhang, X.H., Chen, X.L., Todd, J.D. and Zhang, Y.Z. Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria. Mol. Microbiol. 111 (2019) 1057-1073. [PMID: 30677184]